Tissue-specific responses to TFAM and mtDNA copy number manipulation in prematurely ageing mice
- PMID: 40587199
- PMCID: PMC12208663
- DOI: 10.7554/eLife.104461
Tissue-specific responses to TFAM and mtDNA copy number manipulation in prematurely ageing mice
Abstract
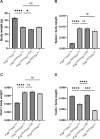
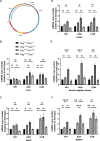
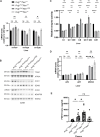
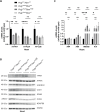
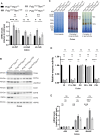
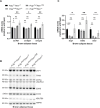
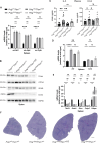
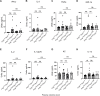
Somatic mitochondrial DNA (mtDNA) mutations are implicated as important drivers of ageing and age-related diseases. Their pathological effect can be counteracted by increasing the absolute amount of wild-type mtDNA via moderately upregulating TFAM, a protein important for mtDNA packaging and expression. However, strong TFAM overexpression can also have detrimental effects as it results in mtDNA hypercompaction and subsequent impairment of mtDNA gene expression. Here, we have experimentally addressed the propensity of moderate TFAM modulation to improve the premature ageing phenotypes of mtDNA mutator mice, carrying random mtDNA mutations. Surprisingly, we detect tissue-specific endogenous compensatory mechanisms acting in mtDNA mutator mice, which largely affect the outcome of TFAM modulation. Accordingly, moderate overexpression of TFAM can have negative and beneficial effects in different tissues of mtDNA mutator mice. We see a similar behavior for TFAM reduction, which improves brown adipocyte tissue homeostasis, while other tissues are unaffected. Our findings highlight that the regulation of mtDNA copy number and gene expression is complex and causes tissue-specific effects that should be considered when modulating TFAM levels. Additionally, we suggest that TFAM is not the sole determinant of mtDNA copy number in situations where oxidative phosphorylation (OXPHOS) is compromised, but other important players must be involved.
Keywords: biochemistry; chemical biology; genetics; genomics; mitochondrial DNA; mouse; mtDNA copy number; mtDNA mutations; tissue specificity.
© 2025, Kremer, Gao et al.
Conflict of interest statement
LK, GG, GR, RF, MM, RW, ÁV, CK No competing interests declared, NL is the inventor of the C5024T mutant mouse licensed to the pharmaceutical industry by the Max Planck Society, and is a scientific founder of Pretzel Therapeutics Inc and owns stock in this company
Figures


Update of
References
-
- Ahlqvist KJ, Leoncini S, Pecorelli A, Wortmann SB, Ahola S, Forsström S, Guerranti R, De Felice C, Smeitink J, Ciccoli L, Hämäläinen RH, Suomalainen A. MtDNA mutagenesis impairs elimination of mitochondria during erythroid maturation leading to enhanced erythrocyte destruction. Nature Communications. 2015;6:1–11. doi: 10.1038/ncomms7494. - DOI - PubMed
-
- Bailey LJ, Cluett TJ, Reyes A, Prolla TA, Poulton J, Leeuwenburgh C, Holt IJ. Mice expressing an error-prone DNA polymerase in mitochondria display elevated replication pausing and chromosomal breakage at fragile sites of mitochondrial DNA. Nucleic Acids Research. 2009;37:2327–2335. doi: 10.1093/nar/gkp091. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

