A Disease-Associated Mutation Impedes PPIA through Allosteric Dynamics Modulation
- PMID: 40587259
- PMCID: PMC12269060
- DOI: 10.1021/acs.biochem.5c00260
A Disease-Associated Mutation Impedes PPIA through Allosteric Dynamics Modulation
Abstract
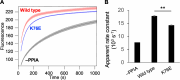
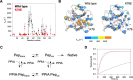
Amyotrophic lateral sclerosis (ALS) is a progressive neurodegenerative disease characterized by motor neuron degeneration. Peptidylprolyl cis-trans isomerase A (PPIA) is a molecular chaperone involved in protein folding, and its dysfunction has been linked to ALS pathogenesis, as proline is recognized as a key residue for maintaining proper folding of ALS-related proteins. A recent study identified a K76E mutation in PPIA in sporadic ALS patients, but its effect on protein function and structure remain unclear. In this study, we used biochemical and biophysical techniques to investigate the structural and functional consequences of the K76E mutation. Our results show that K76E significantly reduces enzyme activity without affecting structure, monodispersity, or substrate recognition. Significant effects of K76E mutation were identified by relaxation dispersion NMR experiments, showing that K76E disrupts key protein dynamics and alters an allosteric network essential for isomerase activity. Corroborated by theoretical kinetic analysis, these dynamics data, revealing the exchange process for K76E to be approximately 1 order of magnitude slower than that of the wild type, explain the reduced cis-trans isomerase activity of the K76E mutant. These findings suggest that the pathogenic effect of K76E arises primarily from impaired protein dynamics rather than direct structural disruption. Our study provides new insights into the molecular mechanisms underlying ALS-associated mutations and their impact on protein function.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

