Light-regulated dual-targeting of NUCLEAR CONTROL OF PEP ACTIVITY establishes photomorphogenesis via interorganellar communication
- PMID: 40587430
- PMCID: PMC12412214
- DOI: 10.1093/plphys/kiaf289
Light-regulated dual-targeting of NUCLEAR CONTROL OF PEP ACTIVITY establishes photomorphogenesis via interorganellar communication
Abstract
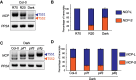
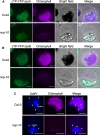
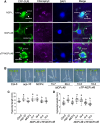
Interorganellar communication is essential for maintaining cellular and organellar functions and adapting to dynamic environmental changes in eukaryotic cells. In angiosperms, light initiates photomorphogenesis, a developmental program characterized by chloroplast biogenesis and inhibition of hypocotyl elongation, through photoreceptors such as the red-/far-red-sensing phytochromes and their downstream signaling pathways. However, the mechanisms underlying nucleus-chloroplast crosstalk during photomorphogenesis remain elusive. Here, we show that light-regulated dual-targeting of NUCLEAR CONTROL OF PEP ACTIVITY (NCP) mediates bidirectional communication between the nucleus and chloroplasts via alternative promoter selection and retrograde translocation. Light promotes transcription from an upstream canonical transcription start site, producing a long NCP (NCP-L) isoform containing an N-terminal chloroplast transit peptide that directs chloroplast localization. In contrast, darkness or low red light conditions favor transcription from a downstream alternative start site, producing a shorter cytoplasmic isoform (NCP-S) that is rapidly degraded via the 26S proteasome. This light-regulated alternative transcription initiation depends on PHYTOCHROME-INTERACTING FACTORS (PIFs), key repressors of photomorphogenesis. Upon chloroplast import, NCP-L is processed into its mature form (NCPm), which promotes assembly and nucleoid localization of the plastid-encoded RNA polymerase (PEP) complex to initiate chloroplast biogenesis. Notably, NCP's nuclear function requires its prior localization to chloroplasts, supporting a model in which NCP mediates chloroplast-to-nucleus retrograde signaling. Consistent with this, NCP promotes stromule formation in Arabidopsis (Arabidopsis thaliana) hypocotyls, linking chloroplast dynamics to phytochrome-dependent nuclear pathways that restrict hypocotyl elongation. Our findings uncover an interorganellar communication mechanism in which light-dependent alternative promoter usage and retrotranslocation regulate photomorphogenesis, integrating nuclear and plastid signals to coordinate organ-specific developmental programs.
© The Author(s) 2025. Published by Oxford University Press on behalf of American Society of Plant Biologists.
Conflict of interest statement
Conflict of interest statement. None declared.
Figures






Similar articles
-
A Holistic Investigation of Arabidopsis Proteomes Altered in Chloroplast Biogenesis and Retrograde Signalling Identifies PsbO as a Key Regulator of Chloroplast Quality Control.Plant Cell Environ. 2025 Aug;48(8):6373-6396. doi: 10.1111/pce.15611. Epub 2025 May 14. Plant Cell Environ. 2025. PMID: 40366233 Free PMC article.
-
Photoexcited CRY1 physically interacts with ATG8 to regulate selective autophagy of HY5 and photomorphogenesis in Arabidopsis.Plant Cell. 2025 Aug 4;37(8):koaf196. doi: 10.1093/plcell/koaf196. Plant Cell. 2025. PMID: 40795357
-
TCP10 acts downstream of light and retrograde signaling pathways to promote Arabidopsis cotyledon de-etiolation in concert with GLK1.New Phytol. 2025 Oct;248(1):265-281. doi: 10.1111/nph.70407. Epub 2025 Jul 24. New Phytol. 2025. PMID: 40704524
-
Plastids in a Pinch: Coordinating Stress and Developmental Responses Through Retrograde Signalling.Plant Cell Environ. 2025 Sep;48(9):6897-6911. doi: 10.1111/pce.15664. Epub 2025 Jun 5. Plant Cell Environ. 2025. PMID: 40474490 Free PMC article. Review.
-
Light signal transduction in plants: insights from phytochrome nuclear translocation and photobody formation.New Phytol. 2025 Sep 12. doi: 10.1111/nph.70572. Online ahead of print. New Phytol. 2025. PMID: 40937522 Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

