Arabidopsis CNL receptor SUT1 confers immunity in hydathodes against the vascular pathogen Xanthomonas campestris pv. campestris
- PMID: 40587573
- PMCID: PMC12233906
- DOI: 10.1371/journal.ppat.1013256
Arabidopsis CNL receptor SUT1 confers immunity in hydathodes against the vascular pathogen Xanthomonas campestris pv. campestris
Abstract
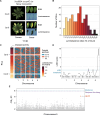
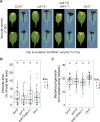
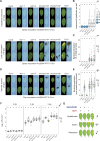
Bacterial plant pathogens exploit natural openings, such as pores or wounds, to enter the plant interior and cause disease. Plants guard these openings through defense mechanisms. However, bacteria from the genus Xanthomonas have specialized in that they enter their host via a special entry point, the hydathode-an organ at the leaf margin involved in xylem sap guttation. Hydathodes can mount an immune response against bacteria, including non-adapted and adapted pathogens like X. campestris pv. campestris (Xcc) that cause vascular disease. Previously, it was shown that the RKS1/ZAR1 immune complex confers vascular resistance against Xcc by recognizing XopAC activity, a type III effector (T3E). However, in absence of XopAC recognition, Arabidopsis Col-0 hydathodes still display resistance against Xcc. Here we mapped the causal gene using an inoculation method that promotes Xcc hydathode entry. Using a population of Recombinant Inbred Lines (RILs) of a cross between a susceptible (Oy-0) and resistant accession (Col-0), a major QTL for Xcc resistance was found on the right arm of Chromosome 5 in Col-0. Combining this result with a genome-wide association analysis yielded a single candidate gene encoding a coiled-coil nucleotide-binding leucine-rich repeat (CNL-type) immune receptor protein called SUPPRESSOR OF TOPP4 1 (SUT1). Expression of SUT1 was confirmed in hydathodes. We reveal that RKS1/ZAR1 and SUT1 confer different levels of Xcc resistance in different tissue types. Both RKS1/ZAR1 and SUT1 are alone sufficient for Xcc resistance in Col-0 hydathodes. However, RKS1/ZAR1 resistance is also effective in tissue types that represent late infection stages, i.e., xylem and mesophyll. In contrast, SUT1 resistance is not effective in the xylem, while weakly additive to RKS1/ZAR1 in the mesophyll. We thus identify a novel R gene, SUT1, that confers Xcc resistance primarily early in the infection during hydathode colonization.
Copyright: © 2025 Taks et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
I have read the journal's policy and the authors of this manuscript have the following competing interests: H.A.v.d.B. is currently an employee of Rijk Zwaan. This work was supported in part by a grant from Rijk Zwaan and Bejo Zaden. Both companies had no role in the design of the study, data collection, and writing of the manuscript.
Figures


Similar articles
-
Delayed Chlorosis in Arabidopsis Ecotype Dijon-G During Bacterial Infection and Dark-Induced Senescence.Physiol Plant. 2025 Jul-Aug;177(4):e70373. doi: 10.1111/ppl.70373. Physiol Plant. 2025. PMID: 40556374 Free PMC article.
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
-
The type-III effectors-based multiplex PCR for detection of Xanthomonas campestris pv. campestris causing black rot disease in crucifer crops.3 Biotech. 2023 Aug;13(8):272. doi: 10.1007/s13205-023-03691-z. Epub 2023 Jul 11. 3 Biotech. 2023. PMID: 37449249 Free PMC article.
-
Infection Assay for Xanthomonas campestris pv. campestris in Arabidopsis thaliana Mimicking Natural Entry via Hydathodes.Methods Mol Biol. 2019;1991:159-185. doi: 10.1007/978-1-4939-9458-8_16. Methods Mol Biol. 2019. PMID: 31041772
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

