Enrichment of low-abundance gene mutation by a combined polymerase and ligase chain reaction
- PMID: 40587690
- PMCID: PMC12212806
- DOI: 10.1097/MD.0000000000042543
Enrichment of low-abundance gene mutation by a combined polymerase and ligase chain reaction
Abstract
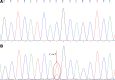
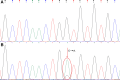
Detection of low-abundance gene mutations or minority alleles in clinical samples is important and challenging in fields of tumor, infectious diseases, noninvasive prenatal diagnosis, and forensic science. The key to solving this problem is the selective enrichment of the low-abundance gene fragments. In this study, a combined polymerase and ligase chain reaction system based on conventional polymerase chain reaction was developed for the first time by introducing a heat-resistant DNA ligase and a pair of ligation primers that target the mutant site. Three EGFR gene mutations (L747_S752 del, G719A, and T790M) were used as the target mutations. Both artificial mixed samples containing 1% of 1 of the 3 EGFR gene mutations and tumor samples were used to evaluate the feasibility of the proposed new method. Sanger sequencing results showed the coexistence of the wild-type and mutant alleles when the amplification product was obtained by the combined polymerase and ligase chain reaction. The novel method based on the combined polymerase and ligase chain reaction has good performance in the detection of the 2 EGFR mutations, as the commercial kit. The novel method could be used to effectively inhibit the amplification of the wild-type gene fragment in the sample and selectively amplify the low-abundance gene fragment with a mutant site, allowing the mutation to be subsequently detected more effectively, accurately, and reliably. By using this novel method, the mutant gene present at an initial content as low as 1% could be effectively amplified and accurately detected.
Keywords: combined polymerase and ligase chain reaction; gene enrichment; heat-resistant DNA ligase; low-abundance gene mutations; minority alleles.
Copyright © 2025 the Author(s). Published by Wolters Kluwer Health, Inc.
Conflict of interest statement
The authors have no conflicts of interest to disclose.
Figures


References
-
- Sidransky D. Emerging molecular markers of cancer. Nat Rev Cancer. 2002;2:210–9. - PubMed
-
- Kobayashi S, Boggon TJ, Dayaram T, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352:786–92. - PubMed
-
- Yoruker EE, Holdenrieder S, Gezer U. Blood-based biomarkers for diagnosis, prognosis and treatment of colorectal cancer. Clin Chim Acta. 2016;455:26–32. - PubMed
MeSH terms
Substances
Grants and funding
- 2019-2-23/Talent Training Project for Health Research of Fujian Province
- 2020J05279/Provincial Natural Science Foundation of Fujian
- YCXZ 19-27/Fujian Maternity and Child Health Hospital Science Startup Foundation
- YCXB 18-04/Doctoral Research Initiation Fund of Fujian Maternity and Child Health Hospital
- 2020Y9140/Fujian Province Science and Technology Innovation Joint Fund Project
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

