CD4+ T cells license Kupffer cells to reverse CD8+ T cell dysfunction induced by hepatocellular priming
- PMID: 40588520
- PMCID: PMC12307227
- DOI: 10.1038/s41590-025-02199-3
CD4+ T cells license Kupffer cells to reverse CD8+ T cell dysfunction induced by hepatocellular priming
Abstract
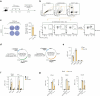
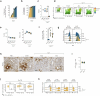
Chronic hepatitis B virus (HBV) infection is marked by dysfunctional HBV-specific CD8+ T cells, and restoring their effector activity is a major therapeutic goal. Here, we generated HBV-specific CD4+ T cell receptor transgenic mice to show that CD4+ effector T cells can prevent and reverse the CD8⁺ T cell dysfunction induced by hepatocellular priming. This rescue enhances antiviral CD8+ T cell function and suppresses viral replication. CD4+ T cell help occurs directly within the liver, independent of secondary lymphoid organs, and requires local antigen recognition. Kupffer cells, rather than dendritic cells, are the critical antigen-presenting platform. CD4+ T cells license Kupffer cells via CD40-CD40L interactions, triggering interleukin (IL)-12 and IL-27 production. IL-12 expands the CD4+ T cell pool, while IL-27 is essential for CD8+ T cell rescue. Exogenous IL-27 similarly restores HBV-specific CD8+ T cell function in mice and in T cells isolated from chronically infected patients. These findings identify IL-27 as a tractable immunotherapeutic target in chronic HBV infection.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: K.R. and T.A.O. are stock owners, and employees at BioNTech. U.S. is stock owner and management board member at BioNTech. P.T.F.K. has acted as a consultant/advisor for Aligos, Assembly Biosciences, Gilead Sciences, GlaxoSmithKline and Bluejay and received educational grants from Aligos, Gilead Sciences and Vir Biotechnology. N.L.B. and A.B. are the founders and hold stocks of T Cell Diagnostics, a biotech company developing T cell tests for viral infection. A.B. is the cofounder of LION TCR, a company developing T cell therapy against viral-related cancers. A.B. consulted and served on the advisory boards of Brii Biosciences, Assembly Biosciences and Gilead Sciences. L.G.G. participated in boards, advisory boards and consultantships for Genenta Science, Epsilen Bio, Gilead Sciences, Antios Therapeutics, Aligos Therapeutics, Medicxi, Chroma Medicine and Ananda Immunotherapies. M.I. participates in advisory boards/consultantships or receives funding from Gilead Sciences, GentiBio, BlueJay Therapeutics, BioNTech, Excision BioTherapeutics, Moderna, GSK and Curie.Bio. The other authors declare no competing interests.
Figures















References
-
- Iannacone, M. & Guidotti, L. G. Immunobiology and pathogenesis of hepatitis B virus infection. Nat. Rev. Immunol.22, 19–32 (2022). - PubMed
-
- Kawashima, K., Andreata, F., Beccaria, C. G. & Iannacone, M. Priming and maintenance of adaptive immunity in the liver. Annu. Rev. Immunol.42, 375–399 (2024). - PubMed
-
- Venzin, V., Beccaria, C. G., Andreata, F., Fumagalli, V. & Iannacone, M. Intrahepatic immunity to hepatitis B virus. J. Hepatol.81, 911–913 (2024). - PubMed
-
- Bertoletti, A. & Ferrari, C. Adaptive immunity in HBV infection. J. Hepatol.64, S71–S83 (2016). - PubMed
-
- Maini, M. K. & Burton, A. R. Restoring, releasing or replacing adaptive immunity in chronic hepatitis B. Nat. Rev. Gastroenterol. Hepatol.16, 662–675 (2019). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

