Sphingosine simultaneously inhibits nuclear import and activates PP2A by binding importins and PPP2R1A
- PMID: 40588551
- PMCID: PMC12361511
- DOI: 10.1038/s44318-025-00490-5
Sphingosine simultaneously inhibits nuclear import and activates PP2A by binding importins and PPP2R1A
Abstract
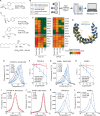
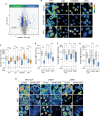
Sphingosine and constrained analogs like FTY720 and SH-BC-893 restrain tumor growth through incompletely defined mechanisms that include protein phosphatase 2A (PP2A) activation. Here we show that these compounds directly bind not only the PP2A scaffolding subunit PPP2R1A, but also the structurally related karyopherins importin-β1 (KPNB1), transportin-1 (TNPO1), importin-5 (IPO5), and importin-7 (IPO7). Binding to sphingosine-like molecules triggers reversible unfolding of these target proteins, resulting in activation of PP2A and inhibition of importins. Although sphingosine engages these proteins, ceramide does not, suggesting that these two endogenous tumor-suppressive sphingolipids work through distinct mechanisms. Simultaneous PP2A activation and importin inhibition reduces nuclear levels of proteins that drive cancer progression and therapeutic resistance such as JUN, YAP, MYC, androgen receptor, hnRNPA1, and NF-κB under conditions where compounds that target PP2A or KPNB1 individually are inactive. These findings provide new insights into sphingolipid biology and highlight a possible path toward cancer therapeutics that could overcome drug resistance.
Keywords: Homeostatic Growth Control; Nuclear Import; Protein Phosphatase 2A; Sphingosine.
© 2025. The Author(s).
Conflict of interest statement
Disclosure and competing interests statement. ALE and SH are inventors on a patent covering SH-BC-893. Other authors declare no competing interests. ALE, PT, and SH are co-corresponding authors. Requests for biological materials should be directed to ALE.
Figures




References
-
- Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, Lindahl E (2015) GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1–2:19–25
-
- Bayliss R, Littlewood T, Stewart M (2000) Structural basis for the interaction between FxFG nucleoporin repeats and importin-beta in nuclear trafficking. Cell 102:99–108 - PubMed
MeSH terms
Substances
Grants and funding
- T32 CA009054/CA/NCI NIH HHS/United States
- 11-23-PDF-03/American Diabetes Association (ADA)
- R01 CA254360/CA/NCI NIH HHS/United States
- BC14027/DOD | MHS | CDMRP | DOD Peer Reviewed Cancer Research Program (PRCRP)
- RSG-11-111-01-CDD/American Cancer Society (ACS)
- R35 GM148350/GM/NIGMS NIH HHS/United States
- P30 CA062203/CA/NCI NIH HHS/United States
- R21 CA178230/CA/NCI NIH HHS/United States
- G-1902-0039/Ono Pharma Foundation
- R35GM148350/HHS | NIH | National Institute of General Medical Sciences (NIGMS)
- 2021R1A6A3A-14039681/National Research Foundation of Korea (NRF)
- R01 GM089919/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Miscellaneous

