The GATE glycoprotein complex enhances human cytomegalovirus entry in endothelial cells
- PMID: 40588592
- PMCID: PMC12312674
- DOI: 10.1038/s41564-025-02025-4
The GATE glycoprotein complex enhances human cytomegalovirus entry in endothelial cells
Abstract
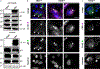
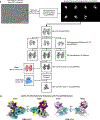
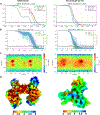
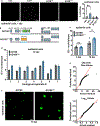
Human cytomegalovirus can cause severe birth defects upon infection in pregnant women and complications in immunocompromised patients. A major challenge for vaccine design is our incomplete understanding of the diverse protein complexes this virus uses to infect cells. In Herpesviridae, glycoproteins H and L (gH and gL) form complexes with other viral proteins that bind receptors to mediate cell-type-specific entry. Here we identify a distinct gH complex that is abundant on human cytomegalovirus virions and enhances infection of endothelial cells. In this complex, gH associates with UL116 and UL141 (an immunoevasin previously known to function intracellularly) but not with gL. We term this the gH-associated tropism and entry (GATE) complex and provide the cryo-electron microscopy structure at ~3.5 Å. The structure shows gH-only scaffolding, UL141-mediated dimerization and a heavily glycosylated UL116 cap. These findings identify a third virion surface complex that promotes cell entry and may represent a new target for vaccines or antiviral therapies.
© 2025. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing interests: M.J.N., M.N.A.S., E.O.S., C.A.B. and J.P.K. are listed as coinventors on US, European and international patent applications related to the identification of GATE-3, US20230293673A1, EP4192494A4 and WO2022032177A1. The other authors declare no competing interests.
Figures











Update of
-
A noncanonical glycoprotein H complex enhances cytomegalovirus entry.bioRxiv [Preprint]. 2024 Oct 13:2024.10.13.617647. doi: 10.1101/2024.10.13.617647. bioRxiv. 2024. Update in: Nat Microbiol. 2025 Jul;10(7):1605-1616. doi: 10.1038/s41564-025-02025-4. PMID: 39416215 Free PMC article. Updated. Preprint.
References
-
- Zuhair M et al. Estimation of the worldwide seroprevalence of cytomegalovirus: A systematic review and meta-analysis. Rev. Med. Virol. 29, e2034 (2019). - PubMed
-
- Chandramouli S et al. Structural basis for potent antibody-mediated neutralization of human cytomegalovirus. Sci Immunol 2, (2017). - PubMed
MeSH terms
Substances
Grants and funding
- R01 AI101423/AI/NIAID NIH HHS/United States
- T32HL155022/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- R01AI139749/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- R01AI116851/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- U19 AI109762/AI/NIAID NIH HHS/United States
- R01 AI116851/AI/NIAID NIH HHS/United States
- P20GM134974/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- R01AI101423/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- T32 HL155022/HL/NHLBI NIH HHS/United States
- U19109762-S1/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- R01 AI139749/AI/NIAID NIH HHS/United States
- P20 GM134974/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

