Epigenetic erosion of H4K20me1 induced by inflammation drives aged stem cell ferroptosis
- PMID: 40588650
- PMCID: PMC12350153
- DOI: 10.1038/s43587-025-00902-5
Epigenetic erosion of H4K20me1 induced by inflammation drives aged stem cell ferroptosis
Abstract
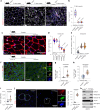
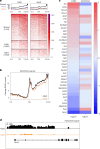
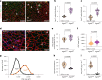
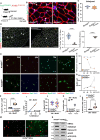
Aging is characterized by a decline in the functionality and number of stem cells across the organism. In this study, we uncovered a mechanism by which systemic inflammation drives muscle stem cell (MuSC) aging through epigenetic erosion. We demonstrate that age-related inflammation decreases monomethylation of H4K20 in MuSCs, disrupting their quiescence and inducing ferroptosis, a form of iron-dependent cell death. Our findings show that inflammatory signals downregulate Kmt5a, the enzyme responsible for depositing H4K20me1, leading to the epigenetic silencing of anti-ferroptosis genes. This results in aberrant iron metabolism, increased reactive oxygen species levels and lipid peroxidation in aged MuSCs. Notably, long-term inhibition of systemic inflammation that is initiated at 12 months of age effectively prevents ferroptosis, preserves MuSC numbers and enhances muscle regeneration and functional recovery. These findings reveal an epigenetic switch that links chronic inflammation to MuSC aging and ferroptosis, offering potential therapeutic strategies for combating age-related muscle degeneration.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures









Update of
-
Epigenetic erosion of H4K20me1 induced by inflammation drives aged stem cell ferroptosis.Res Sq [Preprint]. 2024 Mar 15:rs.3.rs-3937628. doi: 10.21203/rs.3.rs-3937628/v2. Res Sq. 2024. Update in: Nat Aging. 2025 Aug;5(8):1491-1509. doi: 10.1038/s43587-025-00902-5. PMID: 38410478 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

