Computational Insights into Glucose Tolerance and Stimulation in a Family 1 β-glucosidase
- PMID: 40589032
- PMCID: PMC12264956
- DOI: 10.1021/acs.jcim.5c00922
Computational Insights into Glucose Tolerance and Stimulation in a Family 1 β-glucosidase
Abstract
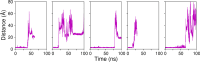
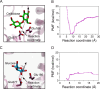
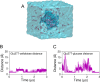
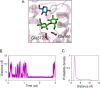
β-Glucosidases catalyze the hydrolysis of cellobiose to glucose during lignocellulosic biomass depolymerization. A significant limitation of many β-glucosidases is product inhibition by glucose, leading to reduced conversion efficiency. However, certain β-glucosidases exhibit tolerance or even stimulation by glucose. The mechanisms underlying this remarkable feature remain poorly elucidated. Here, we employ molecular dynamics simulations to investigate the molecular basis of glucose tolerance and stimulation within the family 1 β-glucosidase from Humicola insolens (HiBgl). Potential of mean force calculations reveal a substantial difference in binding free energies between cellobiose (-12.5 kcal/mol) and glucose (-4.3 kcal/mol) at the HiBgl active site, indicating that the glucose product is a considerably weaker ligand than the cellobiose substrate. These findings are consistent with our observations that HiBgl undergoes conformational changes in its substrate binding site, specifically involving the Trp349 side chain, in the presence of glucose, potentially facilitating glucose expulsion and mitigating product inhibition. Simulations of HiBgl solvated in a 200 mM aqueous glucose environment show that glucose molecules from the bulk solution are capable of penetrating and widening the substrate binding pocket, forming direct interactions with cellobiose in the active site, which may contribute to catalytic stimulation. Additionally, we identify seven distinct secondary glucose binding sites located on the HiBgl surface, spatially distant from the active site, implying a potential role in allosteric regulation. Finally, we demonstrate that glucose at subsite +1 can adopt multiple orientations relative to glucose at subsite -1, a prerequisite for transglycosylation reactions in HiBgl. Our findings elucidate the molecular mechanisms governing HiBgl's glucose tolerance and stimulation, thereby enabling the design of site-directed mutagenesis experiments to improve enzyme efficiency for industrial applications, particularly in biofuel production and oligosaccharide synthesis.
Figures



Similar articles
-
Glucose binding to the gatekeeper region induces the glucose stimulation of β-glucosidase Cba3 from Cellulomonas biazotea.Int J Biol Macromol. 2025 Aug;319(Pt 4):145590. doi: 10.1016/j.ijbiomac.2025.145590. Epub 2025 Jun 29. Int J Biol Macromol. 2025. PMID: 40592427
-
Short-Term Memory Impairment.2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 31424720 Free Books & Documents.
-
Comparison of active site mutations at subsite + 2 of Anoxybacillus ayderensis A9 β-glucosidase for hydrolysis of pNPG and polydatin.BMC Biotechnol. 2025 Jul 1;25(1):52. doi: 10.1186/s12896-025-00984-4. BMC Biotechnol. 2025. PMID: 40598098 Free PMC article.
-
The effect of sample site and collection procedure on identification of SARS-CoV-2 infection.Cochrane Database Syst Rev. 2024 Dec 16;12(12):CD014780. doi: 10.1002/14651858.CD014780. Cochrane Database Syst Rev. 2024. PMID: 39679851 Free PMC article.
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
References
-
- Periyasamy S., Adego A. A., Kumar P. S., Desta G. G., Zelalem T., Karthik V., Isabel J. B., Jayakumar M., Sundramurthy V. P., Rangasamy G.. Influencing Factors and Environmental Feasibility Analysis of Agricultural Waste Preprocessing Routes Towards Biofuel Production – A Review. Biomass Bioenergy. 2024;180:107001. doi: 10.1016/j.biombioe.2023.107001. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

