Qimai Feiluoping Decoction Inhibits EndMT to Alleviate Pulmonary Fibrosis by Reducing PI3K/AKT/mTOR Pathway-Mediated the Restoration of Autophagy
- PMID: 40589610
- PMCID: PMC12208135
- DOI: 10.2147/JIR.S515286
Qimai Feiluoping Decoction Inhibits EndMT to Alleviate Pulmonary Fibrosis by Reducing PI3K/AKT/mTOR Pathway-Mediated the Restoration of Autophagy
Abstract
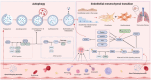
Purpose: Pulmonary Fibrosis (PF) is a severe interstitial lung disease currently lacking effective prevention strategies. Endothelial mesenchymal transition (EndMT), a novel mechanism for fibroblast production, is closely associated with PF. The precise mechanisms underlying the contribution of EndMT-derived fibroblasts to PF, however, remain unclear.
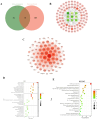
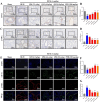
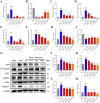
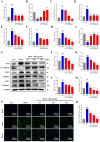
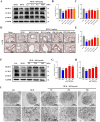
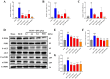
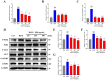
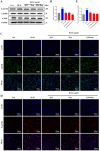
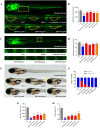
Methods: Using network pharmacology, molecular docking, and molecular dynamics, we identified the key targets and pathways through which Qimai Feiluoping decoction (QM) combats PF. EndMT and autophagy proteins were quantified in bleomycin (BLM) -induced C57BL/6 mice, human umbilical vein endothelial cells (HUVECs), and zebrafish using Western blotting (WB), quantitative real-time polymerase chain reaction (qRT-PCR), immunohistochemistry (IHC), immunofluorescence (IF), Transwell migration assays, and transmission electron microscopy (TEM), revealing the targets and pathways through which QM mitigates PF.
Results: Network pharmacology, molecular docking, and molecular dynamics suggest that QM combats PF by modulating the PI3K/AKT/mTOR pathway. Observations from the study indicated that QM was found to alleviate EndMT by restoring autophagy, primarily through inhibition of the PI3K/AKT/mTOR signaling pathway in both BLM-induced C57 mice and HUVECs. Supporting evidence from zebrafish models demonstrated that QM not only counteracts EndMT but also improves a range of vascular functional disorders and remodeling issues following EndMT.
Conclusion: Our research validates the active compounds, core targets, and signaling pathways through which QM counters PF, providing valuable insights for its therapeutic application in PF management.
Keywords: Qimai Feiluoping decoction; autophagy; endothelial mesenchymal transition; molecular docking; network pharmacology; pulmonary fibrosis.
© 2025 Ma et al.
Conflict of interest statement
The authors report no conflicts of interest in this work.
Figures




References
LinkOut - more resources
Full Text Sources
Miscellaneous

