Environmental Drivers of Genetic Divergence in Two Corals From the Florida Keys
- PMID: 40589611
- PMCID: PMC12206660
- DOI: 10.1111/eva.70126
Environmental Drivers of Genetic Divergence in Two Corals From the Florida Keys
Abstract
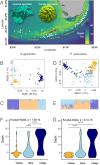
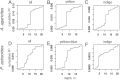
Increasingly frequent marine heatwaves devastate coral reefs around the world, so there is great interest in finding warm-adapted coral populations that could be used as sources for assisted gene flow and restoration. Here, we evaluated the relative power of various environmental factors to explain coral genetic variation, suggestive of differential local adaptation to these factors, across the Florida Keys Reef Tract. We applied a machine learning population genomic method (RDAforest) to two coral species-the mustard hill coral Porites astreoides and the lettuce coral Agaricia agaricites-sampled from 65 sites covering the whole reef tract. Both species comprised three genetically distinct lineages distributed across depths in a remarkably similar way. Within these lineages, there was additional genetic divergence explained by depth, but even more within-lineage variation was cumulatively explained by water chemistry parameters related to nitrogen, phosphorus, silicate, and salinity. Visualizing the predicted environment-associated genetic variation on a geographic map suggests that these associations reflect adaptation to certain aspects of the inshore-offshore environmental gradient, and, to a lesser extent, to difference of Middle and Lower Keys from the rest of the reef tract. Thermal parameters, most notably maximal monthly thermal anomaly, were also consistently identified as putative drivers of genetic divergence, but had a relatively low explanatory power compared to depth and water chemistry. Overall, our results indicate that temperature was not the most important driver of coral genetic divergence in the Florida Keys, and underscore depth and water chemistry as more important environmental factors from the corals' perspective. Our study emphasizes the need for considering a variety of environmental variables, rather than solely focusing on temperature, when predicting how corals may respond to transplantation.
Keywords: 2bRAD; cryptic speciation; polygenic adaptation; random forest; seascape genomics.
© 2025 The Author(s). Evolutionary Applications published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
-
- Aitken, S. , and Whitlock M.. 2013. “Assisted Gene Flow to Facilitate Local Adaptation to Climate Change.” Annual Review of Ecology, Evolution, and Systematics 44: 367–388.
-
- Bairos‐Novak, K. R. , Hoogenboom M. O., van Oppen M. J. H., and Connolly S. R.. 2021. “Coral Adaptation to Climate Change: Meta‐Analysis Reveals High Heritability Across Multiple Traits.” Global Change Biology 27, no. 22: 5694–5710. - PubMed
-
- Baker, A. , and Cunning R.. 2016. “Bulk gDNA Extraction From Coral Samples.” Accessed 2025 February 25. https://www.protocols.io/view/Bulk‐gDNA‐extraction‐from‐coral‐samples‐dy....
-
- Banister, R. B. , and van Woesik R.. 2021. “Ranking 67 Florida Reefs for Survival of Acropora cervicornis Outplants.” Frontiers in Marine Science 8: 672574.
LinkOut - more resources
Full Text Sources

