Therapeutic insights and molecular mechanism linking melatonin signaling and membranous nephropathy (Review)
- PMID: 40589719
- PMCID: PMC12207060
- DOI: 10.3892/br.2025.2017
Therapeutic insights and molecular mechanism linking melatonin signaling and membranous nephropathy (Review)
Abstract
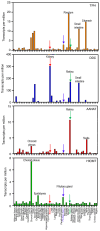
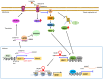
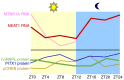
Endogenous melatonin is synthesized at night by specific enzymes and exerts various physiological effects through both melatonin receptor-dependent and -independent pathways. Moreover, exogenous melatonin has been demonstrated to have pleiotropic therapeutic effects on a range of pathological conditions, including renal diseases. The melatonin signaling pathway involves specific enzymes responsible for melatonin synthesis and cellular responses mediated by melatonin, which are evolutionarily conserved in both brain and peripheral tissues. Although the physiological functions of the melatonin-mediated signaling pathway are well-documented across multiple organ systems, its effects on the kidney are less recognized. The present review summarizes the expression levels of melatonin biosynthesis enzymes and melatonin receptors, as well as their roles in renal tissue under pathological conditions such as membranous nephropathy (MN). The present review explores the molecular mechanisms regulating the expression of aryl-alkyl-amine N-acetyl-transferase, nuclear enriched abundant transcript 1 and melatonin receptor 1A (MTNR1A) in renal tubular epithelial cells. Overall, the present review provides new insights into the role of MTNR1A in the pathology, treatment and prevention of MN.
Keywords: AANAT; MN; MTNR1A; NEAT1; melatonin; renal TECs.
Copyright: © 2025 Huang et al.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
Similar articles
-
Melatonin Alleviates Albumin-Induced Tubular Cell Injury by Activating Clock-Controlled Nuclear Enriched Abundant Transcript 1-Mediated Proliferation.ACS Pharmacol Transl Sci. 2024 Oct 10;7(11):3607-3617. doi: 10.1021/acsptsci.4c00495. eCollection 2024 Nov 8. ACS Pharmacol Transl Sci. 2024. PMID: 39539256
-
Melatonin for the promotion of sleep in adults in the intensive care unit.Cochrane Database Syst Rev. 2018 May 10;5(5):CD012455. doi: 10.1002/14651858.CD012455.pub2. Cochrane Database Syst Rev. 2018. PMID: 29746721 Free PMC article.
-
Pharmacotherapies for sleep disturbances in dementia.Cochrane Database Syst Rev. 2016 Nov 16;11(11):CD009178. doi: 10.1002/14651858.CD009178.pub3. Cochrane Database Syst Rev. 2016. Update in: Cochrane Database Syst Rev. 2020 Nov 15;11:CD009178. doi: 10.1002/14651858.CD009178.pub4. PMID: 27851868 Free PMC article. Updated.
-
Melatonin receptor-mediated signaling pathways drive time-dependent physiological recovery from freezing in Perccottus glenii.Fish Physiol Biochem. 2025 Jun 26;51(4):116. doi: 10.1007/s10695-025-01516-9. Fish Physiol Biochem. 2025. PMID: 40569340
-
Health professionals' experience of teamwork education in acute hospital settings: a systematic review of qualitative literature.JBI Database System Rev Implement Rep. 2016 Apr;14(4):96-137. doi: 10.11124/JBISRIR-2016-1843. JBI Database System Rev Implement Rep. 2016. PMID: 27532314
References
Publication types
LinkOut - more resources
Full Text Sources
