Oral bacterial community dynamics during induction of gingival inflammation
- PMID: 40589868
- PMCID: PMC12206739
- DOI: 10.3389/fcimb.2025.1597690
Oral bacterial community dynamics during induction of gingival inflammation
Abstract
Introduction: The human oral cavity is a complex and dynamic microbial ecosystem integral to oral and overall health. While the specific roles of microbial communities in health and disease are not fully understood, dysbiosis of the oral microbiota is, along with inadequate immune fitness, recognized as a key factor driving the onset of inflammatory conditions such as gingivitis. Gingivitis, an early and reversible stage of periodontal disease, involves shifts in microbial composition and diversity. This study aimed to investigate the compositional dynamics of the oral microbiota during the early stages of gingival inflammation, focusing on changes across multiple oral niches and their relationship to clinical outcomes.
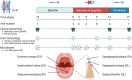
Methods: We conducted an experimental gingivitis intervention study with 41 healthy volunteers. After a two-week baseline period, participants refrained from oral hygiene for two weeks to induce gingivitis, followed by a one-week resolution phase with resumed oral hygiene. Clinical parameters, including plaque and bleeding scores, were monitored at seven time points. Samples from saliva and five oral niches (tongue, keratinized gingiva, supragingival, subgingival, and interproximal dental plaque) were collected and analyzed using 16S rRNA gene sequencing. Multivariate statistical analyses were applied to evaluate microbial dynamics and their associations with clinical outcomes.
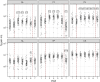
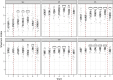
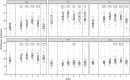
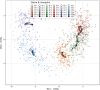
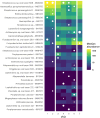
Results: The study revealed pronounced microbial changes, particularly in supragingival plaque, where Leptotrichia and Prevotella increased while Streptococcus decreased. Alpha diversity significantly increased in supragingival plaque, tongue, and saliva during gingivitis induction, highlighting shifts in microbial complexity. Clinical correlations indicated that plaque presence was primarily associated with bacterial load, while gingival bleeding was driven by compositional changes in supragingival plaque and tongue biofilms. These findings suggest that microbial density and composition independently contribute to gingivitis markers.
Conclusion: This study concludes that occurrence of dental plaque and gingival bleeding are independent clinical parameters, linked to bacterial load and composition, respectively. Interactions between multiple niches, especially the tongue, influence clinical outcomes, highlighting a complex, nonlinear dynamic behavior in the oral microbiota. These findings suggest intricate ecological interactions that may approach tipping points, advancing understanding of microbial dynamics during gingival inflammation and informing future strategies for managing gingivitis.
Keywords: 16S rRNA gene sequencing; dysbiosis; gingivitis; inflammation; microbial dynamics; oral microbiota; plaque.
Copyright © 2025 Keijser, van den Broek, van der Wurff, Dulos, Jagers, Kool, Rosema, Nicu, Crielaard, Loos, Brandt and Zaura.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- al-Essa L., Niwa M., Kohno K., Tsurumi K. (1994). A proposal for purification of salivary polymorphonuclear leukocytes by combination of nylon mesh filtration and density-gradient method: a validation by superoxide- and cyclic AMP-generating responses. Life Sci. 55, Pl333–Pl338. doi: 10.1016/0024-3205(94)00773-X - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

