Genetic and Functional Dissection of the NFKB2 Gene: Implications for Milk Fatty Acid Biosynthesis in Dairy Cattle
- PMID: 40590069
- PMCID: PMC12210100
- DOI: 10.1096/fj.202501411R
Genetic and Functional Dissection of the NFKB2 Gene: Implications for Milk Fatty Acid Biosynthesis in Dairy Cattle
Abstract
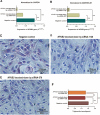
Milk fat, an essential component of bovine milk, significantly impacts human health and dairy industry profitability. Our previous research identified that the NFKB2 gene was a key candidate for milk fatty acids (FAs) in dairy cattle. This study aimed to investigate the role of NFKB2 in regulating milk FAs in Holstein cattle through post-GWAS analysis. Knockdown of NFKB2 using siRNA in bovine mammary epithelial cells (BMECs) significantly reduced triglyceride concentrations, lipid droplet secretion, and the expressions of 17 key genes involved in lipid metabolism pathways, including PI3K-AKT, NF-κB, MAPK, adipocytokine, PPAR, and AMPK signaling pathways. These findings highlighted NFKB2 as a critical regulator of milk FA composition through its interactions with key lipid biosynthesis pathways. Further, this study constructed a new Chinese Holstein population including 1065 cows and performed genotyping and milk FAs measurements in these individuals. By SNP identification and association analysis, SNP g.22889812C/T in the 5' flanking region of NFKB2 was significantly associated with multiple milk FAs, including higher C14:1, C17:0, C18:0, and C14 index levels in allele C group, and higher C6:0, C8:0, C14:0, C16:1, and C16 index levels in allele T group. Using computational predictions, luciferase assays, EMSA and super-shift EMSA, we revealed that SNP g.22889812C/T downregulated NFKB2 transcription activity with allele T selectively binding with transcription factor (TF) ZNF282. This study not only advanced the understanding of genetic mechanisms underlying milk fat production but also identified NFKB2 and its causal mutation g.22889812C/T as promising molecular markers for selective breeding to optimize milk fat traits.
Keywords: EMSA; dairy cattle; fatty acids; knocked‐down of NFKB2; transcriptional activity.
© 2025 The Author(s). The FASEB Journal published by Wiley Periodicals LLC on behalf of Federation of American Societies for Experimental Biology.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
-
- Palombo V., Milanesi M., Sgorlon S., et al., “Genome‐Wide Association Study of Milk Fatty Acid Composition in Italian Simmental and Italian Holstein Cows Using Single Nucleotide Polymorphism Arrays,” Journal of Dairy Science 101 (2018): 11004–11019. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

