Targeting MTAP increases PARP inhibitor susceptibility in triple-negative breast cancer through a feed-forward loop
- PMID: 40590219
- PMCID: PMC12208554
- DOI: 10.1172/JCI188120
Targeting MTAP increases PARP inhibitor susceptibility in triple-negative breast cancer through a feed-forward loop
Abstract
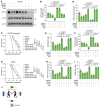
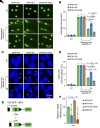
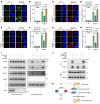
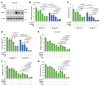
Triple-negative breast cancer (TNBC) represents the most malignant subtype of breast cancer. The clinical application of PARP inhibitors (PARPi) is limited by the low frequency of BRCA1/2 mutations in TNBC. Here, we identified that MTAP deletion sensitized genotoxic agents in our clinical cohort of metastatic TNBC. Further study demonstrated that MTAP deficiency or inhibition rendered TNBC susceptibility to chemotherapeutic agents, particularly PARPi. Mechanistically, targeting MTAP that synergized with PARPi by disrupting the METTL16-MAT2A axis involved in methionine metabolism and depleting in vivo s-adenosylmethionine (SAM) levels. Exhausted SAM in turn impaired PARPi-induced DNA damage repair through attenuation of MRE11 recruitment and end resection by diminishing MRE11 methylation. Notably, brain metastatic TNBC markedly benefited from a lower dose of PARPi and MTAP deficiency/inhibition synergy due to the inherently limited methionine environment in the brain. Collectively, our findings revealed a feed-forward loop between methionine metabolism and DNA repair through SAM, highlighting a therapeutic strategy of PARPi combined with MTAP deficiency/inhibition for TNBC.
Keywords: Amino acid metabolism; Breast cancer; DNA repair; Oncology; Therapeutics.
Figures
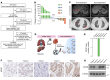
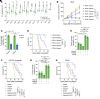
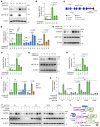
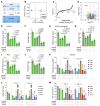


Comment in
- A triple-punch approach: methionine restriction enhances combination inhibitors in brain metastatic triple-negative breast cancer doi: 10.1172/JCI193171
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

