A haploinsufficiency restoration strategy corrects neurobehavioral deficits in Nf1+/- mice
- PMID: 40590220
- PMCID: PMC12208548
- DOI: 10.1172/JCI188932
A haploinsufficiency restoration strategy corrects neurobehavioral deficits in Nf1+/- mice
Abstract
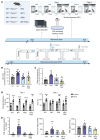
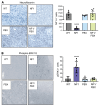
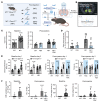
Neurofibromatosis type 1 (NF1) is a genetic disorder caused by mutations of the NF1 tumor suppressor gene resulting in the loss of function of neurofibromin, a GTPase-activating protein (GAP) for Ras. While the malignant manifestations of NF1 are associated with loss of heterozygosity of the residual WT allele, the nonmalignant neurodevelopmental sequelae, including autism spectrum disorder (ASD) and/or attention deficit hyperactivity disorder (ADHD) are prevalent morbidities that occur in the setting of neurofibromin haploinsufficiency. We reasoned that augmenting endogenous levels of WT neurofibromin could serve as a potential therapeutic strategy to correct the neurodevelopmental manifestations of NF1. Here, we used a combination of genetic screening and genetically engineered murine models to identify a role for the F-box protein FBXW11 as a regulator of neurofibromin degradation. Disruption of Fbxw11, through germline mutation or targeted genetic manipulation in the nucleus accumbens, increased neurofibromin levels, suppressed Ras-dependent ERK phosphorylation, and corrected social learning deficits and impulsive behaviors in male Nf1+/- mice. Our results demonstrate that preventing the degradation of neurofibromin is a feasible and effective approach to ameliorate the neurodevelopmental phenotypes in a haploinsufficient disease model.
Keywords: Genetic diseases; Genetics; Neurodevelopment; Neuroscience; Ubiquitin-proteosome system.
Figures





References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

