Neuropilin-2 functions as a coinhibitory receptor to regulate antigen-induced inflammation and allograft rejection
- PMID: 40590224
- PMCID: PMC12208552
- DOI: 10.1172/JCI172218
Neuropilin-2 functions as a coinhibitory receptor to regulate antigen-induced inflammation and allograft rejection
Abstract
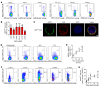
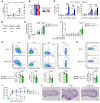
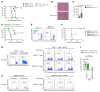
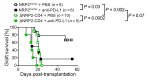
Coinhibitory receptors function as central modulators of the immune response to resolve T effector activation and/or to sustain immune homeostasis. Here, using humanized SCID mice, we found that neuropilin-2 (NRP2) is inducible on late effector and exhausted subsets of human CD4+ T cells and that it is coexpressed with established coinhibitory molecules including PD-1, CTLA4, TIGIT, LAG3, and TIM3. In murine models, we also found that NRP2 is expressed on effector memory CD4+ T cells with an exhausted phenotype and that it functions as a key coinhibitory molecule. Knockout (KO) of NRP2 resulted in hyperactive CD4+ T cell responses and enhanced inflammation in delayed-type hypersensitivity and transplantation models. After cardiac transplantation, allograft rejection and graft failure were accelerated in global as well as CD4+ T cell-specific KO recipients, and enhanced alloimmunity was dependent on NRP2 expression on CD4+ T effectors but not on CD4+Foxp3+ Tregs. Also, KO Tregs were found to be as efficient as WT cells in the suppression of effector responses in vitro and in vivo. These collective findings identify NRP2 as a potentially novel coinhibitory receptor and demonstrate that its expression on CD4+ T effector cells is of great functional importance in immunity.
Keywords: Cellular immune response; Immunology; Organ transplantation; T cells; Transplantation.
Conflict of interest statement
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

