N6-methyladenosine methylation: a novel key to unlocking mental disorders
- PMID: 40590504
- PMCID: PMC12309383
- DOI: 10.1093/ijnp/pyaf044
N6-methyladenosine methylation: a novel key to unlocking mental disorders
Abstract
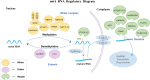
More than 100 types of RNA modifications have been identified in mammalian cells, among which N6-methyladenosine (m6A) is the most prevalent. This reversible and dynamic modification involves methyltransferases, demethylases, and reader proteins. Aberrant expression of m6A-related regulatory proteins in the nervous system significantly impacts neuronal physiology, contributing to mental disorders such as depression, autism spectrum disorder, and schizophrenia. This review summarizes the role of m6A methylation in the pathogenesis of mental disorders and highlights its potential as a biomarker and therapeutic target, providing a comprehensive reference for future research and clinical interventions.
Keywords: learning and memory; m6A methylation; mental disorders; substance addiction.
© The Author(s) 2025. Published by Oxford University Press on behalf of the CINP.
Conflict of interest statement
The authors declare that there is no conflict of interest.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

