Functional proteomic analysis of Streptomyces sp. F-3 reveals its potential to effectively degrade waste-yeast
- PMID: 40590931
- PMCID: PMC12214041
- DOI: 10.1007/s00253-025-13541-y
Functional proteomic analysis of Streptomyces sp. F-3 reveals its potential to effectively degrade waste-yeast
Abstract
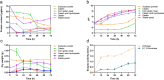
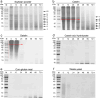
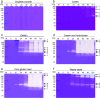
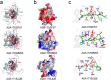
Streptomyces are renowned in pharmaceutical and medical fields for their ability to produce antibiotics and other bioactive secondary metabolites. In order to reduce industrial production costs, it is crucial to find suitable and cheaper raw materials as carbon and nitrogen sources for microbial growth processes. This study investigated the substrate preference of Streptomyces sp. F-3 using functional proteomic analysis. Streptomyces sp. F-3 exhibited varying degradation and utilization rates for different nitrogen source. The results indicated that the strain F-3 could not efficiently degrade intact globular proteins, but preferred to degrade peptone or protein hydrolysate, especially for waste-yeast. The strain F-3 could utilize waste-yeast to grow rapidly and produced a large amount of extracellular protein. The substrate-binding patterns of three S8 proteases secreted by Streptomyces sp. F-3 determined the nitrogen source degradation preference of the strain. In addition, the strain F-3 could secrete large amounts of β-glucanase and chitinase to utilize cell wall polysaccharides. Thus, waste-yeast, rich in peptone, β-glucan, and chitin, could be the superior substrate for culturing Streptomyces. This study not only broadens the application scenarios for waste-yeast, but also provides valuable insights for rapid and cost-effective industrial microbial cultivation. KEY POINTS: The substrate preference of Streptomyces sp. F-3 was analyzed by integrative omics. Structural omics revealed the hydrolysis specificity of S8 proteases from F-3. Waste-yeast served as the superior substrate for culturing Streptomyces.
Keywords: Streptomyces sp. F-3; Functional proteomic analysis; Proteases; Substrate preference; Waste-yeast.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethical approval: This article does not contain any studies with human participants or animals performed by any of the authors. Conflict of interest: The authors declare no competing interests.
Figures


References
-
- Al-Dhabi NA, Esmail GA, Ghilan AKM, Arasu MV, Duraipandiyan V, Ponmurugan K (2020) Characterization and fermentation optimization of novel thermo stable alkaline protease from Streptomyces sp. Al-Dhabi-82 from the Saudi Arabian environment for eco-friendly and industrial applications. King Saud Univ Sci 32(1):1258–1264. 10.1016/j.jksus.2019.11.011
-
- Bartlett GJ, Porter CT, Borkakoti N, Thornton JM (2002) Analysis of catalytic residues in enzyme active sites. J Mol Biol 324(1):105–121 10.1016/s0022-2836(02)01036-7 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

