Activity-based probes for dynamic characterisation of polysaccharide-degrading enzymes
- PMID: 40591799
- PMCID: PMC12312396
- DOI: 10.1042/BCJ20253060
Activity-based probes for dynamic characterisation of polysaccharide-degrading enzymes
Abstract
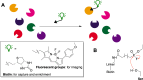
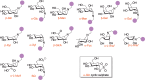
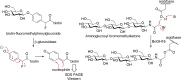
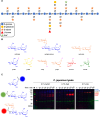
Carbohydrate-active enzymes play essential roles in polysaccharide degradation, yet their biochemical characterisation remains challenging - especially in the face of rapidly expanding genomic and structural data. Standard annotations often overlook critical properties such as expression patterns, enzyme stability and substrate specificity, which are key to understanding function in biological and industrial contexts. Activity-based probes (ABPs) offer a direct solution by enabling selective detection of active enzymes within complex systems. This review focuses on ABPs for retaining glycosidases, tracing their development from early applications in medical diagnostics to emerging uses in biomass degradation. We examine recent advances in scaffold design - including fluorosugars, epoxides, aziridines and cyclic sulphates - and their utility in enzyme profiling, inhibitor discovery and biotechnology. Current ABPs remain limited: they cannot yet target inverting enzymes and other classes lacking nucleophilic residues, a gap that may be bridged through computational modelling and AI-guided probe development. Looking forward, integration of ABPs with enzyme engineering and design holds promise for unlocking new classes of biocatalysts tailored for industrial and biomedical use.
Keywords: activity-based probes; biomass conversion; carbohydrate-active enzymes; enzyme inhibitors; glycobiology.
© 2025 The Author(s).
Conflict of interest statement
The authors declare that there are no competing interests associated with the manuscript.
Figures



References
-
- Thaler M., Ofman T.P., Kok K., Heming J.J.A., Moran E., Pickles I., et al. Epi-cyclophellitol cyclosulfate, a mechanism-based endoplasmic reticulum α-glucosidase ii inhibitor, blocks replication of sars-cov-2 and other coronaviruses. ACS Cent. Sci. 2024;10:1594–1608. doi: 10.1021/acscentsci.4c00506. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

