Comprehensive genetic profiling of sensorineural hearing loss using an integrative diagnostic approach
- PMID: 40592345
- PMCID: PMC12281402
- DOI: 10.1016/j.xcrm.2025.102206
Comprehensive genetic profiling of sensorineural hearing loss using an integrative diagnostic approach
Abstract
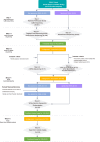
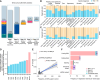
Despite the advent of next-generation sequencing, diagnosing genetic disorders remains challenging. We perform comprehensive genomic profiling of 394 families (752 individuals) with sensorineural hearing loss (SNHL) using a systematic multi-tiered approach, from single-gene analysis to whole-genome sequencing (WGS), complemented by functional assays and bioinformatic analysis. Our strategy achieves a cumulative diagnostic yield of 55.6% (219 families), with automated WGS analysis identifying pathogenic variants in an additional 20 families, primarily structural variants. Comparative analysis reveals higher frequencies of single pathogenic alleles in recessive genes within our cohort compared to controls. Subsequent analysis, including in silico predictions and in vitro validation, identifies three deep intronic pathogenic variants on opposite alleles. These findings demonstrate the value of comprehensive genomic analysis in resolving undiagnosed cases. Finally, we map the genome-phenome landscape of SNHL at the level of inner ear function. Our results highlight WGS as a transformative tool for precision medicine in genetic diseases.
Keywords: genotype-phenotype correlations; molecular diagnostics; precision medicine; rare genetic disorder; sensorineural hearing loss; stepwise genomic approach; whole-genome sequencing.
Copyright © 2025 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests Y.S.J. is the founder of Inocras, Inc., a genome analysis and interpretation company. Y.S.J., J.-Y.K., S.P., and Sangmoon Lee hold stocks or stock options in Inocras, Inc.
Figures




References
-
- Martin A.R., Williams E., Foulger R.E., Leigh S., Daugherty L.C., Niblock O., Leong I.U.S., Smith K.R., Gerasimenko O., Haraldsdottir E., et al. PanelApp crowdsources expert knowledge to establish consensus diagnostic gene panels. Nat. Genet. 2019;51:1560–1565. doi: 10.1038/s41588-019-0528-2. - DOI - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

