Mutant p53 induces SH3BGRL expression to promote cell engulfment
- PMID: 40592808
- PMCID: PMC12218370
- DOI: 10.1038/s41420-025-02582-x
Mutant p53 induces SH3BGRL expression to promote cell engulfment
Abstract
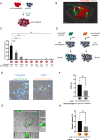
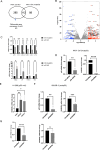
Previously, we identified that mutant p53 expression in cancer cells promotes engulfment of neighbouring cancer cells to form cell-in-cell (CIC) structures. This process gave mutant p53 cells an advantage in tumour formation in mouse xenograft experiments. TP53 can be found mutated at nearly every amino acid in cancers and mutant p53 expression is associated with various GOF (Gain-of-function) processes, including cancer cell invasion, metastasis, stemness and drug resistance. In the current manuscript, we identified SH3BGRL (Src homology 3 binding glutamate rich protein like) as a mutant p53-regulated gene and investigated to what extent SH3BGRL expression and cell engulfment are responsible for mutant p53-dependent anchorage-independent growth and chemoresistance. We demonstrate that mutant p53 expression drives cell engulfment in which the mutant p53 host cell moves in the direction of the target internal cell to form CIC structures. This is therefore more reminiscent of cell engulfment rather than cell entosis, in which cells invade into host cells. Using NGS (Next Generation Sequencing), we identified novel target genes of mutant p53 and demonstrate that cell engulfment requires SH3BGRL expression. We generated mutant p53 and p53 KO cell lines that stably overexpressed SH3BGRL and determined that SH3BGRL promotes etoposide resistance in mutant p53 cells and anchorage-independent growth independent of mutant p53 expression. Through FACS sorting of pure cell engulfing (CIC) populations, we could also show that engulfing cells have an enhanced etoposide resistance. These data suggest that SH3BGRL and cell engulfment are required for certain GOFs of mutant p53.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests. Ethics approval: All animal experiments were approved by the AWERB (application number: BIOL-2022-04-21T09_50_20-pkkp47) of Durham University and under project licence number: PP8383164. Animal experiments are reported under ARIVE guidelines. For patient studies, ethics approval was not required as only publicly available database were used including the p53 database: https://tp53.cancer.gov/about and several patient datasets accessible via Xenabrowser: https://xenabrowser.net .
Figures




Similar articles
-
Behavioral interventions to reduce risk for sexual transmission of HIV among men who have sex with men.Cochrane Database Syst Rev. 2008 Jul 16;(3):CD001230. doi: 10.1002/14651858.CD001230.pub2. Cochrane Database Syst Rev. 2008. PMID: 18646068
-
Interventions for promoting habitual exercise in people living with and beyond cancer.Cochrane Database Syst Rev. 2018 Sep 19;9(9):CD010192. doi: 10.1002/14651858.CD010192.pub3. Cochrane Database Syst Rev. 2018. PMID: 30229557 Free PMC article.
-
Antidepressants for pain management in adults with chronic pain: a network meta-analysis.Health Technol Assess. 2024 Oct;28(62):1-155. doi: 10.3310/MKRT2948. Health Technol Assess. 2024. PMID: 39367772 Free PMC article.
-
Maternal and neonatal outcomes of elective induction of labor.Evid Rep Technol Assess (Full Rep). 2009 Mar;(176):1-257. Evid Rep Technol Assess (Full Rep). 2009. PMID: 19408970 Free PMC article.
-
Atypical antipsychotics for disruptive behaviour disorders in children and youths.Cochrane Database Syst Rev. 2017 Aug 9;8(8):CD008559. doi: 10.1002/14651858.CD008559.pub3. Cochrane Database Syst Rev. 2017. PMID: 28791693 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

