Machine learning reveals genes impacting oxidative stress resistance across yeasts
- PMID: 40592811
- PMCID: PMC12215403
- DOI: 10.1038/s41467-025-60189-3
Machine learning reveals genes impacting oxidative stress resistance across yeasts
Abstract
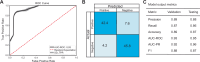
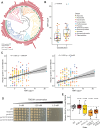
Reactive oxygen species (ROS) are highly reactive molecules encountered by yeasts during routine metabolism and during interactions with other organisms, including host infection. Here, we characterize the variation in resistance to the ROS-inducing compound tert-butyl hydroperoxide across the ancient yeast subphylum Saccharomycotina and use machine learning (ML) to identify gene families whose sizes are predictive of ROS resistance. The most predictive features are enriched in gene families related to cell wall organization and include two reductase gene families. We estimate the quantitative contributions of features to each species' classification to guide experimental validation and show that overexpression of the old yellow enzyme (OYE) reductase increases ROS resistance in Kluyveromyces lactis, while Saccharomyces cerevisiae mutants lacking multiple mannosyltransferase-encoding genes are hypersensitive to ROS. Altogether, this work provides a framework for how ML can uncover genetic mechanisms underlying trait variation across diverse species and inform trait manipulation for clinical and biotechnological applications.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: A.R. is a scientific consultant for LifeMine Therapeutics, Inc. The other authors declare no competing interests.
Figures




Similar articles
-
Transcriptomic insights into Candida albicans adaptation to an anaerobic environment.Microbiol Spectr. 2025 Jul;13(7):e0302424. doi: 10.1128/spectrum.03024-24. Epub 2025 May 22. Microbiol Spectr. 2025. PMID: 40401963 Free PMC article.
-
Cells resist starvation through a nutrient stress splice switch.Nucleic Acids Res. 2025 Jun 20;53(12):gkaf525. doi: 10.1093/nar/gkaf525. Nucleic Acids Res. 2025. PMID: 40550515 Free PMC article.
-
Combating Reactive Oxygen Species (ROS) with Antioxidant Supramolecular Polymers.ACS Appl Mater Interfaces. 2025 Jun 18;17(24):35275-35287. doi: 10.1021/acsami.5c06967. Epub 2025 Jun 3. ACS Appl Mater Interfaces. 2025. PMID: 40462549 Free PMC article.
-
EORTC guidelines for the use of erythropoietic proteins in anaemic patients with cancer: 2006 update.Eur J Cancer. 2007 Jan;43(2):258-70. doi: 10.1016/j.ejca.2006.10.014. Epub 2006 Dec 19. Eur J Cancer. 2007. PMID: 17182241
-
Towards the next generation of species delimitation methods: an overview of machine learning applications.Mol Phylogenet Evol. 2025 Sep;210:108368. doi: 10.1016/j.ympev.2025.108368. Epub 2025 May 16. Mol Phylogenet Evol. 2025. PMID: 40348350 Review.
References
-
- Kurtzman, C. P., Fell, J. W. & Boekhout, T. The Yeasts: A Taxonomic Study, 5, Elsevier (Elsevier, 2011).
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources

