Identification of immunogenic and cross-reactive chikungunya virus epitopes for CD4+ T cells in chronic chikungunya disease
- PMID: 40592820
- PMCID: PMC12218217
- DOI: 10.1038/s41467-025-60862-7
Identification of immunogenic and cross-reactive chikungunya virus epitopes for CD4+ T cells in chronic chikungunya disease
Abstract
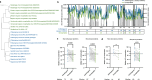
Chikungunya virus (CHIKV), a mosquito-borne alphavirus, causes acute febrile illness that can progress into chronic arthritis-like disease (CHIKVD) in humans. CD4+ T cells have important functions in CHIKV infection, yet the CHIKV target proteins for these CD4 + T cells are poorly characterized. Here, by stimulating PBMCs collected from individuals with chronic CHIKVD with peptides spanning the entire CHIKV proteome, we provide a comprehensive landscape of CHIKV CD4+ T cell epitopes. We identify three immunodominant regions and associated core motifs in CHIKV E1, nsP1 and CP proteins. In addition, by in silico assessment of the sequence conservation of CHIKV proteome with closely related alphaviruses, we define CHIKV epitopes conserved across arthritogenic and encephalitic viruses. Overall, our work describes CD4+ T cell targets of CHIKV in humans, thereby assisting in studying the functions of CD4+ T cells in CHIKV pathogenesis and vaccine design.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: D.W is a consultant for Moderna. A.S. is a consultant for Alcimed, Arcturus, Darwin Health, Desna Therapeutics, EmerVax, Gilead Sciences, Guggenheim Securities, Link University and RiverVest Venture Partners. LJI has filed for patent protection for various aspects of T cell epitope and vaccine design work. The remaining authors declare no conflicts of interest.
Figures
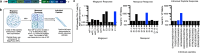


Similar articles
-
Chikungunya virus-specific CD4+ T cells are associated with chronic chikungunya viral arthritic disease in humans.Cell Rep Med. 2025 May 20;6(5):102134. doi: 10.1016/j.xcrm.2025.102134. Cell Rep Med. 2025. PMID: 40398392 Free PMC article.
-
Chikungunya in a pediatric cohort: Asymptomatic infection, seroconversion, and chronicity rates.PLoS Negl Trop Dis. 2025 Jul 16;19(7):e0013254. doi: 10.1371/journal.pntd.0013254. eCollection 2025 Jul. PLoS Negl Trop Dis. 2025. PMID: 40668874 Free PMC article.
-
Humoral and cellular immune response to a single dose of a novel bivalent recombinant adenovirus-vector vaccine against West Nile virus and chikungunya virus in mice.Virol J. 2025 Jul 25;22(1):256. doi: 10.1186/s12985-025-02878-5. Virol J. 2025. PMID: 40713773 Free PMC article.
-
Evidence of previous but not current transmission of chikungunya virus in southern and central Vietnam: Results from a systematic review and a seroprevalence study in four locations.PLoS Negl Trop Dis. 2018 Feb 9;12(2):e0006246. doi: 10.1371/journal.pntd.0006246. eCollection 2018 Feb. PLoS Negl Trop Dis. 2018. PMID: 29425199 Free PMC article.
-
Exploiting the chikungunya virus capsid protein: a focused target for antiviral therapeutic development.Arch Virol. 2025 May 27;170(7):141. doi: 10.1007/s00705-025-06325-2. Arch Virol. 2025. PMID: 40423856 Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

