New molecular components of high and low affinity iron import systems in Drosophila
- PMID: 40592826
- PMCID: PMC12218971
- DOI: 10.1038/s41467-025-60758-6
New molecular components of high and low affinity iron import systems in Drosophila
Abstract
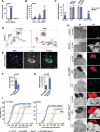
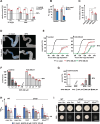
The high abundance and molecular versatility of iron have led to its universal presence in biological systems, yet its absorption is exceptionally challenging. Animals and yeasts use divalent metal transporters to import iron, but yeasts also employ the multicopper oxidase Fet3p for high-affinity iron uptake when iron-starved. Using long-term iron depletion in Drosophila, we identified four components involved in iron absorption: Multicopper oxidase-4 (Mco4), a Fet3p ortholog, is essential for surviving iron starvation, whereas the cytochrome b561 enzymes Fire (Ferric Iron Reductase) and Fire-like, as well as cytochrome b5 protein Firewood, are required for iron absorption under normal conditions. This study reports the presence of a high-affinity iron uptake system in an animal, a cytochrome b5 electron donor for ferric iron reduction, and intestinal ferric reductases, and provides a valuable resource for further exploration of genes involved in iron homeostasis, transport, and absorption.
© 2025. Crown.
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




Similar articles
-
Converging Roles of the Metal Transporter SMF11 and the Ferric Reductase FRE1 in Iron Homeostasis of Candida albicans.Mol Microbiol. 2024 Dec;122(6):879-895. doi: 10.1111/mmi.15326. Epub 2024 Nov 11. Mol Microbiol. 2024. PMID: 39529282
-
EORTC guidelines for the use of erythropoietic proteins in anaemic patients with cancer: 2006 update.Eur J Cancer. 2007 Jan;43(2):258-70. doi: 10.1016/j.ejca.2006.10.014. Epub 2006 Dec 19. Eur J Cancer. 2007. PMID: 17182241
-
Interventions to reduce harm from continued tobacco use.Cochrane Database Syst Rev. 2016 Oct 13;10(10):CD005231. doi: 10.1002/14651858.CD005231.pub3. Cochrane Database Syst Rev. 2016. PMID: 27734465 Free PMC article.
-
Starvation Induces Upregulation of Monocarboxylate Transport in Glial Cells at the Drosophila Blood-Brain Barrier.Glia. 2025 Aug;73(8):1608-1626. doi: 10.1002/glia.70021. Epub 2025 Apr 16. Glia. 2025. PMID: 40241296
-
Fortification of salt with iron and iodine versus fortification of salt with iodine alone for improving iron and iodine status.Cochrane Database Syst Rev. 2022 Apr 21;4(4):CD013463. doi: 10.1002/14651858.CD013463.pub2. Cochrane Database Syst Rev. 2022. PMID: 35446435 Free PMC article.
References
-
- Lill, R. Function and biogenesis of iron-sulphur proteins. Nature460, 831–838 (2009). - PubMed
MeSH terms
Substances
Grants and funding
- P30 GM133894/GM/NIGMS NIH HHS/United States
- PS 169102/Gouvernement du Canada | Canadian Institutes of Health Research (Instituts de Recherche en Santé du Canada)
- RGPIN-2018-04357/Canadian Network for Research and Innovation in Machining Technology, Natural Sciences and Engineering Research Council of Canada (NSERC Canadian Network for Research and Innovation in Machining Technology)
LinkOut - more resources
Full Text Sources
Medical

