Autoregulation of the real-time kinetics of the human mitochondrial replicative helicase
- PMID: 40592829
- PMCID: PMC12214949
- DOI: 10.1038/s41467-025-60289-0
Autoregulation of the real-time kinetics of the human mitochondrial replicative helicase
Abstract
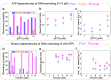
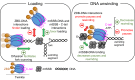
The human mitochondrial helicase Twinkle is essential for mitochondrial DNA (mtDNA) replication and integrity. Using biochemical and single-molecule techniques, we investigated Twinkle's real-time kinetics, including DNA loading, unwinding, and rewinding, and their regulation by its N-terminal Zinc-binding domain (ZBD), C-terminal tail, and mitochondrial SSB protein (mtSSB). Our results indicate that Twinkle rapidly scans dsDNA to locate the fork, where specific interactions halt diffusion. During unwinding, ZBD-DNA interactions and C-terminal tail control of ATPase activity downregulate kinetics, slowing down the helicase. Binding of mtSSB to DNA likely outcompetes ZBD-DNA interactions, alleviating the downregulatory effects of this domain. Furthermore, we show that ZBD-DNA interactions and ATP binding also regulate rewinding kinetics following helicase stalling. Our findings reveal that ZBD and C-terminal tail play a major role in regulation of Twinkle´s real-time kinetics. Their interplay constitutes an auto-regulatory mechanism that may be relevant for coordinating the mtDNA maintenance activities of the helicase.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



References
-
- Spelbrink, J. N. et al. Human mitochondrial DNA deletions associated with mutations in the gene encoding Twinkle, a phage T7 gene 4-like protein localized in mitochondria. Nat. Genet.28, 223–231 (2001). - PubMed
MeSH terms
Substances
Grants and funding
- PID2020-120258GB-I00/Ministry of Economy and Competitiveness | Agencia Estatal de Investigación (Spanish Agencia Estatal de Investigación)
- HR24-00604/"la Caixa" Foundation (Caixa Foundation)
- R15 GM139104/GM/NIGMS NIH HHS/United States
- PGC2018-099341-B-I00/Ministry of Economy and Competitiveness | Agencia Estatal de Investigación (Spanish Agencia Estatal de Investigación)
- PID2021-126755NB-I00/Ministry of Economy and Competitiveness | Agencia Estatal de Investigación (Spanish Agencia Estatal de Investigación)
LinkOut - more resources
Full Text Sources

