Rewiring of 3D chromatin topology orchestrates transcriptional reprogramming in muscle fiber-type specification and transformation
- PMID: 40592831
- PMCID: PMC12219728
- DOI: 10.1038/s41467-025-60866-3
Rewiring of 3D chromatin topology orchestrates transcriptional reprogramming in muscle fiber-type specification and transformation
Abstract
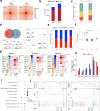
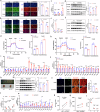
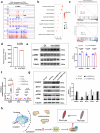
The composition of muscle fibers, characterized by distinct contractile and metabolic properties, significantly influences meat quality and glucose homeostasis. However, the mechanisms by which three-dimensional (3D) genome topology integrates with epigenetic states to regulate muscle fiber specification and transformation remain poorly understood. Here, we present an integrative analysis of the transcriptome, epigenome, and 3D genome architecture in the slow-twitch glycolytic extensor digitorum longus (EDL) and fast-twitch oxidative soleus (SOL) muscles of the pig (Sus scrofa). Global remodeling of enhancer-promoter (E-P) interactions emerged as a central driver of transcriptional reprogramming associated with muscle contraction and glucose metabolism. We identified tissue-specific super-enhancers (SEs) that regulate muscle fiber-type specification through cooperation of chromatin looping and transcription factors such as KLF5. Notably, the SE-driven activation of STARD7 facilitated the transformation of glycolytic fibers into oxidative fibers by mitigating reactive oxygen species levels and suppressing ERK MAPK signaling. This study elucidates the principles of 3D genome organization in the epigenetic regulation of muscle fiber specification and transformation, providing a foundation for novel therapeutic strategies targeting metabolic disorders and enhancing meat quality.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




References
-
- Picard, B., Lefaucheur, L., Berri, C. & Duclos, M. J. Muscle fibre ontogenesis in farm animal species. Reprod. Nutr. Dev.42, 415–431 (2002). - PubMed
-
- Schiaffino, S. & Reggiani, C. Fiber types in mammalian skeletal muscles. Physiol. Rev.91, 1447–1531 (2011). - PubMed
-
- Oberbach, A. et al. Altered fiber distribution and fiber-specific glycolytic and oxidative enzyme activity in skeletal muscle of patients with type 2 diabetes. Diabetes Care29, 895–900 (2006). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

