N6-methyladenosine modification of MEF2A weakens cetuximab sensitivity in colorectal cancer via PD-L1/SOX12 axis
- PMID: 40592832
- PMCID: PMC12219012
- DOI: 10.1038/s41420-025-02577-8
N6-methyladenosine modification of MEF2A weakens cetuximab sensitivity in colorectal cancer via PD-L1/SOX12 axis
Abstract
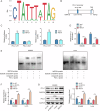
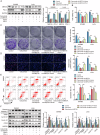
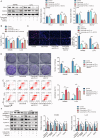
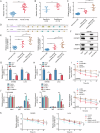
Colorectal cancer (CRC) treatment is still a challenge due to chemoresistance. We explored MEF2A function and underlying mechanism on cetuximab sensitivity in CRC. In this study, cancer tissues and adjacent non-cancerous samples were harvested from CRC patients. Cell viability, proliferation and apoptosis in CRC cells were tested by CCK-8, EdU, colony formation, and flow cytometry assays. The binding of MEF2A on the PD-L1 promoter was validated using luciferase reporter assay, CHIP, and EMSA, while the relationship of PD-L1 and SOX12 mRNA, as well as RBM15/IGF2BP1 and MEF2A mRNA, was verified by RIP, RNA pull-down, or FISH combined with immunofluorescence. m6A modification level of MEF2A mRNA was assayed by MeRIP. The expressions of key genes and proteins, including MEF2A, PD-L1, SOX12, RBM15, IGF2BP1, apoptosis- and cell cycle-related proteins, were determined with RT-qPCR, western blot, or immunohistochemistry. In vivo function of MEF2A was validated by establishing a xenograft nude mice model. The results showed that MEF2A was increased in CRC cells and tissues, while it was higher in cetuximab-resistant CRC tissues. Silencing MEF2A improved the sensitivity of cetuximab in CRC cells and xenograft mice. MEF2A binds to PD-L1 promoter to transcriptionally upregulate PD-L1 expression. Increased cetuximab sensitivity was observed in PD-L1 knockout (KO) CRC cells. PD-L1 overexpression reversed the enhanced cetuximab sensitivity induced by MEF2A knockdown. PD-L1 binds to SOX12 mRNA to stabilize its expression. PD-L1 knockdown augmented cetuximab sensitivity, which was overturned by SOX12 overexpression. The m6A modification mediated by RBM15/IGF2BP1 upregulated MEF2A expression in cetuximab-resistant CRC tissues. In conclusion, m6A-modified MEF2A alleviated cetuximab sensitivity in CRC via PD-L1/SOX12 mRNA axis, indicating that MEF2A might function as a promising therapeutic target against cetuximab-resistant CRC.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests. Ethics approval and consent to participate: The present research was approved by the Ethics Committee of Soochow University, and written consent was gained from all individuals. Animal experiments had been approved by the Animal Care and Use Committee of Soochow University.
Figures




References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

