Accounting for population structure and data quality in demographic inference with linkage disequilibrium methods
- PMID: 40592851
- PMCID: PMC12218012
- DOI: 10.1038/s41467-025-61378-w
Accounting for population structure and data quality in demographic inference with linkage disequilibrium methods
Abstract
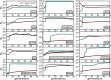
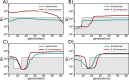
Linkage disequilibrium methods for demographic inference usually rely on panmictic population models. However, the structure of natural populations is generally complex and the quality of the genotyping data is often suboptimal. We present two software tools that implement theoretical developments to estimate the effective population size (Ne): GONE2, for inferring recent changes in Ne when a genetic map is available, and currentNe2, which estimates contemporary Ne even in the absence of genetic maps. These tools operate on SNP data from a single sample of individuals, and provide insights into population structure, including the FST index, migration rate, and subpopulation number. GONE2 can also handle haploid data, genotyping errors, and low sequencing depth data. Results from simulations and laboratory populations of Drosophila melanogaster validated the tools in different demographic scenarios, and analysis were extended to populations of several species. These results highlight that ignoring population subdivision often leads to Ne underestimation.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


Similar articles
-
Estimating Recent and Historical Effective Population Size of Marine and Freshwater Sticklebacks.Mol Ecol. 2025 Jul;34(13):e17825. doi: 10.1111/mec.17825. Epub 2025 Jun 8. Mol Ecol. 2025. PMID: 40485047 Free PMC article.
-
Effectiveness and cost-effectiveness of computer and other electronic aids for smoking cessation: a systematic review and network meta-analysis.Health Technol Assess. 2012;16(38):1-205, iii-v. doi: 10.3310/hta16380. Health Technol Assess. 2012. PMID: 23046909
-
The clinical effectiveness and cost-effectiveness of enzyme replacement therapy for Gaucher's disease: a systematic review.Health Technol Assess. 2006 Jul;10(24):iii-iv, ix-136. doi: 10.3310/hta10240. Health Technol Assess. 2006. PMID: 16796930
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
-
A rapid and systematic review of the clinical effectiveness and cost-effectiveness of paclitaxel, docetaxel, gemcitabine and vinorelbine in non-small-cell lung cancer.Health Technol Assess. 2001;5(32):1-195. doi: 10.3310/hta5320. Health Technol Assess. 2001. PMID: 12065068
References
-
- Frankham, R., Ballou, J. D. & Briscoe, D. A. Introduction to Conservation Genetics 2nd ed (Cambridge University Press, Cambridge, U.K., 2010).
-
- Caballero, A. Quantitative Genetics (Cambridge University Press, 2020).
-
- Tallmon, D. A., Koyuk, A., Luikart, G. & Beaumont, M. A. ONeSAMP: a program to estimate effective population size using approximate Bayesian computation. Mol. Ecol. Resour.8, 299–301 (2008). - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

