Generating combinatorial diversity via engineered V(D)J-like recombination in Saccharomyces cerevisiae
- PMID: 40592886
- PMCID: PMC12216023
- DOI: 10.1038/s41467-025-61206-1
Generating combinatorial diversity via engineered V(D)J-like recombination in Saccharomyces cerevisiae
Abstract
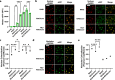
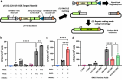
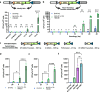
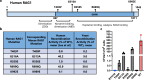
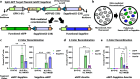
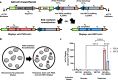
V(D)J recombination is integral to the development of antibody diversity and proceeds through a complex DNA cleavage and repair process mediated by several proteins, including recombination-activating genes 1 and 2, RAG1 and RAG2. V(D)J recombination occurs in all jawed vertebrates but is absent from evolutionarily distant relatives, including the yeast Saccharomyces cerevisiae. As yeast grow quickly and are a platform for antibody display, engineering yeast to undergo V(D)J recombination could expand their applicability for studying antibody development. Therefore, in this work we incorporate RAG1 and RAG2 into yeast and characterize the resulting recombination ability using a split antibiotic resistance assay, demonstrating successful homology-assisted formation of coding joints. By pursuing a variety of strategies, we increase the rate of homology-assisted recombination by over 7000-fold, with the best rates approaching 1% recombination after four days. We further show that our platform can assay the severity of several disease-causing RAG1 mutations. Finally, we use our engineered yeast to simultaneously generate up to three unique fluorescent proteins or two distinct antibody fragments starting from an array of nonfunctional gene fragments, which we believe to be the first-ever generation of genetic and phenotypic diversity solely using random recombination of preexisting DNA in a non-vertebrate cell.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The Authors declare the following competing interests: J.B. and A.P.C. have filed a patent related to this work. The other authors declare no competing interests.
Figures

References
-
- Schatz, D. G. et al. The Mechanism, Regulation and Evolution of V(D)J Recombination. in Molecular Biology of B Cells (Third Edition) Chapter 2, (eds. Honjo, T., Reth, M., Radbruch, A., Alt, F. & Martin, A.) 13–57 (Academic Press, London, 2024).
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

