MEN: leveraging explainable multimodal encoding network for precision prediction of CYP450 inhibitors
- PMID: 40592952
- PMCID: PMC12214809
- DOI: 10.1038/s41598-025-04982-6
MEN: leveraging explainable multimodal encoding network for precision prediction of CYP450 inhibitors
Abstract
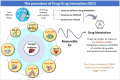
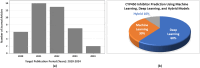
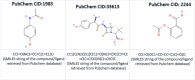
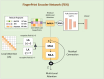
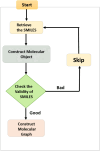
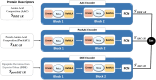
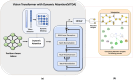
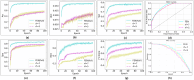
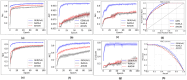
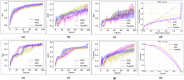
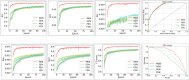
Drug-drug interactions (DDIs) present serious risks in clinical settings, especially for patients who are prescribed multiple medications. A major factor contributing to these interactions is the inhibition of cytochrome P450 (CYP450) enzymes, which are vital for drug metabolism. As a result, reliably identifying compounds that may inhibit CYP450 enzymes is a key step in drug development. However, existing machine learning (ML) methods often fall short in terms of prediction accuracy and biological interpretability. To address this challenge, we introduce a Multimodal Encoder Network (MEN) aimed at improving the prediction of CYP450 inhibitors. This model combines three types of molecular data (chemical fingerprints, molecular graphs, and protein sequences) by applying specialized encoders tailored to each format. Specifically, the Fingerprint Encoder Network (FEN) processes molecular fingerprints, the Graph Encoder Network (GEN) extracts structural features from graph-based representations, and the Protein Encoder Network (PEN) captures sequential patterns from protein sequences. By integrating these diverse data types, MEN can extract complementary information that enhances predictive performance. The encoded outputs from FEN, GEN, and PEN are fused to build a comprehensive feature representation. An explainable AI (XAI) module is incorporated into the model to support biological interpretation, using visualization techniques such as heatmaps. The model was trained and validated using two datasets: chemical structures in SMILES format from PubChem and protein sequences of five CYP450 isoforms (1A2, 2C9, 2C19, 2D6, and 3A4) obtained from the Protein Data Bank (PDB). MEN achieved an average accuracy of 93.7% across all isoforms. The individual encoders performed with accuracies of 80.8% (FEN), 82.3% (GEN), and 81.5% (PEN). Additional performance results include an AUC of 98.5%, sensitivity of 95.9%, specificity of 97.2%, precision of 80.6%, F1-score of 83.4%, and a Matthews correlation coefficient (MCC) of 88.2%. All data and code are available at https://github.com/GracedAbena/MEN-Leveraging-Explainable-Multimodal-Encoding-Network .
Keywords: Attentive hierarchical graph isomorphism network (AHGIN); Cytochrome P450; Inhibitor; Interpretable artificial intelligence (IAI); Residual multi local attention (ReMLA); Self-attention.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures













Similar articles
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
-
DDINet: Drug-drug interaction prediction network based on multi-molecular fingerprint features and multi-head attention centered weighted autoencoder.J Bioinform Comput Biol. 2025 Feb;23(1):2550003. doi: 10.1142/S0219720025500039. J Bioinform Comput Biol. 2025. PMID: 40169368
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
-
Antidepressants for pain management in adults with chronic pain: a network meta-analysis.Health Technol Assess. 2024 Oct;28(62):1-155. doi: 10.3310/MKRT2948. Health Technol Assess. 2024. PMID: 39367772 Free PMC article.
-
Leveraging a foundation model zoo for cell similarity search in oncological microscopy across devices.Front Oncol. 2025 Jun 18;15:1480384. doi: 10.3389/fonc.2025.1480384. eCollection 2025. Front Oncol. 2025. PMID: 40606969 Free PMC article.
References
-
- Li, X. et al. Prediction of human cytochrome P450 inhibition using a multitask deep autoencoder neural network. Mol. Pharm.15(10), 4336–4345. 10.1021/acs.molpharmaceut.8b00110 (2018). - PubMed
-
- Arimoto, R. Computational models for predicting interactions with cytochrome p450 enzyme. Curr. Top. Med. Chem.6(15), 1609–1618 (2006). - PubMed
-
- Ingelman-Sundberg, M. Pharmacogenetics of cytochrome P450 and its applications in drug therapy: the past, present and future. Trends Pharmacol. Sci.25(4), 193–200. 10.1016/j.tips.2004.02.007 (2004). - PubMed
-
- Zhou, S.-F., Liu, J.-P. & Chowbay, B. Polymorphism of human cytochrome P450 enzymes and its clinical impact. Drug Metab. Rev.41(2), 89–295. 10.1080/03602530902843483 (2009). - PubMed
MeSH terms
Substances
Grants and funding
- RS-2023-00256517/This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT)
- (IITP-2025-RS-2024-00437191)/This work was supported by the IITP(Institute of Information & Communications Technology Planning & Evaluation)-ITRC(Information Technology Research Center) grant funded by the Korea government(Ministry of Science and ICT)
LinkOut - more resources
Full Text Sources
Research Materials

