Functional characterisation of components in two Plasmodium falciparum Cullin-RING-Ligase complexes
- PMID: 40592953
- PMCID: PMC12218279
- DOI: 10.1038/s41598-025-05342-0
Functional characterisation of components in two Plasmodium falciparum Cullin-RING-Ligase complexes
Abstract
Ubiquitination is the key eukaryotic post-translational modification that governs protein degradation, localisation, and activity which is mediated by a concerted enzyme cascade. The largest superfamily of these enzymes include the Cullin-RING-Ligase (CRL) complexes. Plasmodium falciparum, the causative agent of the most severe form of malaria in humans, encodes the critical proteins required for ubiquitination, but we do not yet understand the function of this pathway. Here the P. falciparum CRL complexes were characterised to reveal an essential but minimal repertoire controlled by two Cullin scaffolds. A PfCullin1-linked CRL complex, recruiting a single substrate receptor, was identified as being required for parasite inner-membrane biogenesis and DNA replication. A second CRL complex functioning through a PfCullin4 scaffold was identified that utilised a previously unidentified adaptor protein and receptors to support DNA replication. These results show that the P. falciparum CRL complexes are essential in both nuclear maintenance and membrane integrity.
Keywords: P. falciparum; Cullin-ring-ligase; E3 ligase; Malaria; Ubiquitination.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests. Ethics statement: Use of human blood and serum was approved by the Walter and Eliza Hall Institute of Medical Research Human Ethics committee under approval number 19-05VIC-13. Human blood and serum were provided by the Australian Red Cross Blood Service who obtained informed consent from donors and/or their legal guardian(s). All methods were carried out in accordance with relevant guidelines and regulations as approved by the Walter and Eliza Hall Institute of Medical Research Institute Ethics Committee.
Figures
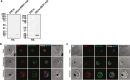


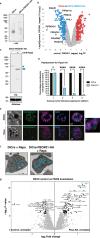
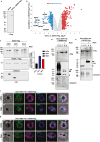
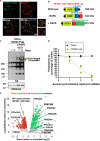
References
-
- WHO. World malaria report 2023. Geneva: World Health Organization. Licence: CC BY-NC-SA 3.0 IGO. (2023).
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

