The genomic comparison between autochthonous and cosmopolitan cows reveals structural variants involved in environmental adaptation
- PMID: 40593057
- PMCID: PMC12215704
- DOI: 10.1038/s41598-025-07165-5
The genomic comparison between autochthonous and cosmopolitan cows reveals structural variants involved in environmental adaptation
Abstract
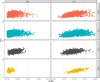
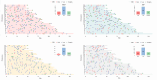
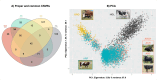
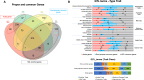
Copy Number Variants (CNVs) are structural variants affecting genetic diversity and phenotypic variability of populations. Different authors underlined the relevance of CNV in relation to the adaptation to environmental conditions (e.g., altitude, harsh farming environment). Aosta cattle (Aosta Red Pied - ARP; Aosta Black Pied/Chestnut - ABC and Mixed Chestnut-Héren - ACH) farmed in the Aosta Valley, and the Oropa Red Pied (ORO), farmed in the Piedmont region, are autochthonous dual-purpose breeds well adapted to the natural Alpine environment. In contrast, the Holstein (HOL) breed is a specialized dairy breed raised in intensive farming systems, representing an artificial environment. The aim of this study is to use CNVs to characterize these breeds and explore the relationship between structural genomic variability and adaptation to mountain farming systems (natural environment) vs. intensive farming systems (artificial environment). Using the GeneSeek Genomic Profiler Bovine 100K data, a total of 160,798 CNVs were identified across 5,610 individuals. Principal Component Analysis (PCA) using CNV Regions (CNVRs) revealed that Aosta breeds clustered into two separate groups, with one smaller cluster including part of ORO cows, while the Holstein formed a distinct cluster. These results suggest that CNVs may act as markers of adaptive selection, influencing both Aosta and ORO breeds, though to a different extent compared to the intensively farmed HOL breed. A total of 526 CNVRs were identified in at least 2% of the samples. Annotated genes and overlapping QTL were functionally associated with production, functional traits, and health-related characteristics. VST analysis revealed candidate genes linked to environmental adaptation, reproduction, and metabolic efficiency in Aosta, ORO, and HOL cattle. Key findings include TCF12 and SRGAP1 deletions in Aosta, suggesting trade-offs between muscle growth and endurance, while ELF2 and ARID5B gains in Holstein indicate aptitude for milk protein synthesis and feed efficiency. Additionally, reproductive genes (RGS3, GSE1, MARCH10) showed distinct selection pressures between Dual-purpose and Holstein breeds, reflecting adaptation to different production systems.
Keywords: Aosta breed; Copy number variant (CNV); Dual-purpose breed; Environmental adaptation; Holstein.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests. Ethical approval: This study used genotyping data provided by ANABORAVA and made available from GENORIP project. Hence, no ethical approval was needed.
Figures

Similar articles
-
Uncovering the architecture of production-driven introgression in Cinisara cattle breed.BMC Genom Data. 2025 Jul 11;26(1):47. doi: 10.1186/s12863-025-01337-y. BMC Genom Data. 2025. PMID: 40640709 Free PMC article.
-
Analysis of genomic selection characteristics of local cattle breeds in Gansu.BMC Genomics. 2025 Jul 1;26(1):574. doi: 10.1186/s12864-025-11753-0. BMC Genomics. 2025. PMID: 40597588 Free PMC article.
-
Multi-tool copy number detection highlights common body size-associated variants in miniature pig breeds from different geographical regions.BMC Genomics. 2025 Mar 22;26(1):285. doi: 10.1186/s12864-025-11446-8. BMC Genomics. 2025. PMID: 40121435 Free PMC article.
-
Advances in the genetic characterization of guinea fowl in Africa: a comprehensive overview of the current status, progress of genome genotyping, and future perspectives.Poult Sci. 2025 May 28;104(9):105363. doi: 10.1016/j.psj.2025.105363. Online ahead of print. Poult Sci. 2025. PMID: 40482533 Free PMC article. Review.
-
A meta-analysis of genome-wide association studies to identify candidate genes associated with feed efficiency traits in pigs.J Anim Sci. 2025 Jan 4;103:skaf010. doi: 10.1093/jas/skaf010. J Anim Sci. 2025. PMID: 39847436 Free PMC article.
References
-
- Meek, M. H. et al. Understanding local adaptation to prepare populations for climate change. Bioscience73, 36–47 (2023).
-
- Hoffmann, I. Adaptation to climate change–exploring the potential of locally adapted breeds. Animal7, 346–362 (2013). - PubMed
-
- Castillo-Salas, C. A. et al. Molecular markers for thermo-tolerance are associated with reproductive and physiological traits in pelibuey Ewes Raised in a semiarid environment. J. Therm. Biol.112, 103475 (2023). - PubMed
Publication types
MeSH terms
Grants and funding
- CUP_J72C21000650005/''Le razze bovine a duplice attitudine: un modello alternativo di zootecnia eco-sostenibile - DUALBREEDING FASE 2" - PSRN Programma di Sviluppo Rurale Nazionale 2014/2020 - Sottomisura 10.2 - PERIODO 2020-2023 - funded by the Ministero dell'Agricoltura della Sovranità Alimentare e delle Foreste with economic support of European Agricultural Fund for Rural Development
- G53D23004040006/Prin 2022 "GENOmic Breeding VALue Estimation in a Native Alpine Cattle Breed" grant - Finanziato dall'Unione europea - Next Generation EU
LinkOut - more resources
Full Text Sources
Miscellaneous

