Evaluation of normalisation strategies for qPCR data obtained from canine gastrointestinal tissues with different pathologies
- PMID: 40593152
- PMCID: PMC12217775
- DOI: 10.1038/s41598-025-06657-8
Evaluation of normalisation strategies for qPCR data obtained from canine gastrointestinal tissues with different pathologies
Abstract
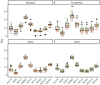
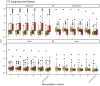
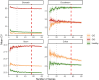
Quantitative real-time PCR (qPCR) is widely used to quantify gene expression at the mRNA level and confirm RNAseq results. Normalisation is a critical process used to minimise technical variability introduced during sample processing. To achieve this, reference genes (RGs), known for their stable expression across conditions, are commonly used as a baseline for accurate comparison. In canine intestinal tissue, there are no published studies validating RGs or other normalisation strategies. This study aimed to identify the best normalisation method in qPCR data from intestinal tissue biopsies of healthy dogs and in dogs suffering from gastrointestinal disease. RNA later-preserved tissue from healthy dogs and dogs with different gastrointestinal pathologies was used for RNA isolation and subsequent qPCR. Ninety-six genes were profiled using a qPCR high-throughput platform. Eleven RGs were included in the investigation. The RGs were ranked based on their stability using two standard methods (GeNorm and NormFinder). Normalisation including one to five of the most stable RGs and the global mean (GM) of the expression of all tested genes as an alternative normalisation method were investigated. The global mean expression was the best-performing normalisation method. Three RGs (RPS5, RPL8 and HMBS) were suitably stable RGs for normalising qPCR data when profiling small sets of genes in canine gastrointestinal tissue with different pathology. Furthermore, in our experimental set-up, with multiple tissues under different conditions, the implementation of the GM method is advisable when a set greater than 55 genes is profiled.
Keywords: CV; Chronic inflammatory enteropathy; Dog; Gastrointestinal cancer; Gastrointestinal tract; Global mean; Normalisation; Reference genes; qPCR.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures
References
-
- Farrell, R. E. in In RNA Methodologies (Sixth Edition). 135–162 (eds Farrell, R. E.) (Academic, 2023).
-
- Raso, A. & Biassoni, R. Quantitative Real-Time PCR (Springer, New York, 2020).
MeSH terms
LinkOut - more resources
Full Text Sources

