High-resolution structure of the heat-stable form-IAq RuBisCO from the thermophilic purple sulfur bacterium Thermochromatium tepidum
- PMID: 40593175
- PMCID: PMC12216891
- DOI: 10.1038/s41598-025-07081-8
High-resolution structure of the heat-stable form-IAq RuBisCO from the thermophilic purple sulfur bacterium Thermochromatium tepidum
Abstract
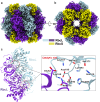
Ribulose 1,5-bisphosphate carboxylase/oxygenase (RuBisCO) catalyzes the initial carbon fixation reaction in the Calvin-Benson-Bassham cycle. Among the many forms of RuBisCOs, form-I-a protein complex containing 8 large and 8 small subunits-is the most common, representing over 90% of all known RuBisCOs. Although many form-I RuBisCO structures have been determined, no structure has been reported for a form-IAq RuBisCO. Here, we detail the structure of the heat-stable form-IAq RuBisCO from the thermophilic and anaerobic purple bacterium Thermochromatium (Tch.) tepidum at 1.55 Å resolution. The overall structure of the Tch. tepidum form-IAq RuBisCO resembles both a form-IAc RuBisCO from a chemolithotrophic sulfur bacterium and a synthetic form-I RuBisCO reconstructed from ancestral sequences. However, the Tch. tepidum enzyme shows significantly greater interactions between adjacent small subunits through their extended N-terminal domains that contain a characteristic six-residue insertion unique to form-IAq RuBisCOs. Structural differences of Tch. tepidum RuBisCO from its mesophilic relative Allochromatium vinosum, and key substitutions on the hydrophilic surface of the small subunits suggests the mechanisms of its enhanced thermostability. Our structure represents the first structure of a form-IAq RuBisCO, providing fresh clues for unraveling the evolutionary history of RuBisCO and new details for how this key enzyme remains active at elevated temperatures.
Keywords: Thermochromatium tepidum; Cryo-EM; Photosynthesis; RuBisCO.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures




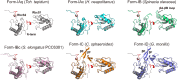
Similar articles
-
Chemoautotrophy in subzero environments and the potential for cold-adapted Rubisco.Appl Environ Microbiol. 2025 Jun 18;91(6):e0060425. doi: 10.1128/aem.00604-25. Epub 2025 May 30. Appl Environ Microbiol. 2025. PMID: 40444981 Free PMC article.
-
Replacement of large subunit N terminus enabled biogenesis of different plant Rubiscos in E. coli.Plant Biotechnol J. 2025 Aug;23(8):3382-3391. doi: 10.1111/pbi.70162. Epub 2025 May 29. Plant Biotechnol J. 2025. PMID: 40443069 Free PMC article.
-
Complete genome of the thermophilic purple sulfur Bacterium Thermochromatium tepidum compared to Allochromatium vinosum and other Chromatiaceae.Photosynth Res. 2022 Jan;151(1):125-142. doi: 10.1007/s11120-021-00870-y. Epub 2021 Oct 20. Photosynth Res. 2022. PMID: 34669148
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2017 Dec 22;12(12):CD011535. doi: 10.1002/14651858.CD011535.pub2. Cochrane Database Syst Rev. 2017. Update in: Cochrane Database Syst Rev. 2020 Jan 9;1:CD011535. doi: 10.1002/14651858.CD011535.pub3. PMID: 29271481 Free PMC article. Updated.
References
-
- Field, C. B., Behrenfeld, M. J., Randerson, J. T. & Falkowski, P. Primary production of the biosphere integrating terrestrial and oceanic components. Science28, 237–240 (1998). - PubMed
-
- Tabita, F. R., Satagopan, S., Hanson, T. E., Kreel, N. E. & Scott, S. S. Distinct form I, II, III, and IV Rubisco proteins from the three kingdoms of life provide clues about Rubisco evolution and structure/function relationships. J. Exp. Bot.59, 1515–1524 (2007). - PubMed
-
- Badger, M. R. & Bek, E. J. Multiple Rubisco forms in proteobacteria: their functional significance in relation to CO2 acquisition by the CBB cycle. J. Exp. Bot.59, 1525–1541 (2008). - PubMed
-
- Newman, J. & Gutteridge, S. Structure of an effector-induced inactivated state of ribulose 1,5-bisphosphate carboxylase Oxygenase the binary complex between enzyme and xylulose 1,5-bisphosphate. Structure2, 495–502 (1994). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

