PKIB facilitates bladder cancer proliferation and metastasis through mediation of HSP27 phosphorylation by PKA
- PMID: 40593489
- PMCID: PMC12219054
- DOI: 10.1038/s41419-025-07814-7
PKIB facilitates bladder cancer proliferation and metastasis through mediation of HSP27 phosphorylation by PKA
Abstract
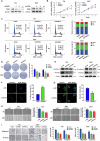
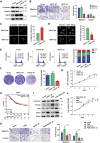
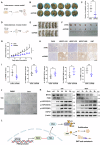
Cyclic AMP-dependent protein kinase A (PKA) is recognized for its pivotal involvement in various cancer types, with Protein Kinase Inhibitor Beta (PKIB) serving as an endogenous inhibitor that curtails PKA activity. Despite the documented escalation of PKIB expression in several malignancies, a comprehensive understanding of its precise mechanistic implications in human cancers remains elusive. This investigation is centered on bladder cancer (BLCA), unveiling an augmented expression of PKIB concomitant with heightened BLCA cell proliferation, migration, and invasion in vitro and augmented tumorigenic potential in an in vivo model. Mechanistically, PKIB disrupts PKA kinase activity, thereby resulting in diminished phosphorylation of the substrate target protein HSP27 at serine 15, 78, and 82. Additionally, the transcription factor MYCN exhibits an affinity for the PKIB promoter, leading to its enhanced expression in the context of BLCA. These findings reveal the oncogenic proclivity of PKIB and introduce a novel signalling pathway in BLCA, providing valuable insights into potential therapeutic targets for precise intervention.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests. Ethical approval and consent to participate: Human BLCA tissues were collected after informed consent was obtained from all patients. The study protocol was conducted in accordance with the Declaration of Helsinki and has been approved by the Academic and Ethics Advisory Committee of the Second Affiliated Hospital of Soochow University (Approval No. [JD-LK-2019-104-01]). Animal experiments were conducted according to guidelines approved by the Animal Care and Use Committee of Soochow University. All animal experiments were approved by the Animal Care and Use Committee of Soochow University.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

