Echs1-mediated histone crotonylation facilitates zygotic genome activation and expression of repetitive elements in early mammalian embryos
- PMID: 40593492
- PMCID: PMC12219887
- DOI: 10.1038/s41467-025-60565-z
Echs1-mediated histone crotonylation facilitates zygotic genome activation and expression of repetitive elements in early mammalian embryos
Abstract
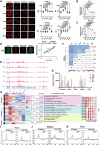
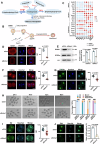
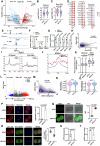
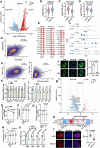
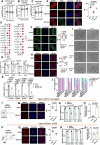
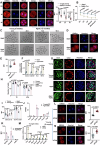
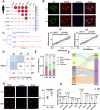
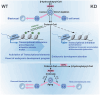
Histone crotonylation, a conserved post-translational histone modification, plays a crucial role in transcriptional regulation. However, its function in early embryonic development remains largely unexplored. Here, we perform genome-wide mapping of histone crotonylation in mouse and human early embryos. Our analysis reveals that histone crotonylation is highly enriched at promoter regions and exhibits distinct dynamic patterns throughout embryogenesis. Notably, strong histone crotonylation signals are observed at the mouse 2-cell and human 4-to-8-cell stages, coinciding with zygotic genome activation. In mice, Echs1 knockdown in oocytes, which suppresses histone crotonylation, results in developmental arrest at the 2-cell stage. Further investigation demonstrates that reduced histone crotonylation impairs transcriptional activity at zygotic genome activation genes, retrotransposon elements, and ribosomal DNA loci. Moreover, early embryos from aged female mice exhibit significantly diminished histone crotonylation, while supplementation with exogenous sodium crotonate enhances blastocyst formation. Collectively, our findings establish histone crotonylation as a key regulatory mechanism in early mammalian embryogenesis by facilitating transcriptional activation of zygotic genome activation genes and repetitive elements.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures
References
-
- Zhang, B. et al. Widespread enhancer dememorization and promoter priming during parental-to-zygotic transition. Mol. cell72, 673–686.e676 (2018). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

