DNA polymerase α/primase extraction from chromatin by VCP/p97 restricts ATR activation during unperturbed DNA replication
- PMID: 40593507
- PMCID: PMC12219636
- DOI: 10.1038/s41467-025-60077-w
DNA polymerase α/primase extraction from chromatin by VCP/p97 restricts ATR activation during unperturbed DNA replication
Abstract
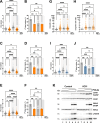
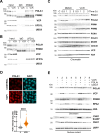
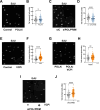
The replication stress response is an essential pathway that deals with the obstacles that halt the progression of DNA replication forks even during an unperturbed S phase. Basal activation of the ATR and CHK1 kinases prevents the premature firing of origins of replication during S phase, avoiding the activation of an excessive number of replication forks and the appearance of genomic instability. However, the mechanisms that regulate ATR activation in the unperturbed S phase have not been fully determined. Here we present evidence that the AAA ATPase VCP/p97 regulates the presence of the DNA polymerase α/Primase complex (POLA/PRIM) on chromatin, thus limiting its activity and hampering the subsequent activation of ATR by TOPBP1. As a consequence, inhibiting VCP/p97 activates ATR and CHK1 and leads to a cell cycle arrest in G2/M. We propose that the priming activity of POLA/PRIM in the lagging strand is one of the determinants of the basal activation of ATR during an unperturbed S phase and VCP/p97 limits this activation through the extraction of POLA/PRIM from chromatin.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

