Epitenon-derived progenitors drive fibrosis and regeneration after flexor tendon injury in a spatially-dependent manner
- PMID: 40593535
- PMCID: PMC12219553
- DOI: 10.1038/s41467-025-60704-6
Epitenon-derived progenitors drive fibrosis and regeneration after flexor tendon injury in a spatially-dependent manner
Abstract
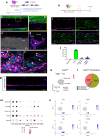
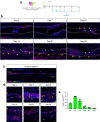
Flexor tendon injuries are common and heal poorly owing to both the deposition of function-limiting peritendinous scar tissue and insufficient healing of the tendon itself. Therapeutic options are limited due to a lack of understanding of the cell populations that contribute to these processes. Here, we identified the epitenon as a major source of cells that contribute to both peritendinous fibrosis and regenerative tendon healing following acute tendon injury. Using a combination of genetic lineage tracing and single cell RNA-sequencing (scRNA-seq), we profiled the behavior and contributions of each cell fate to the healing process in a spatio-temporal manner. Integrated scRNA-seq analysis of mouse healing with human peritendinous scar tissue revealed remarkable transcriptional similarity between mouse epitenon-derived cells and fibroblasts present in human peritendinous scar tissue, which was further validated by immunofluorescent staining for conserved markers. Finally, ablation of pro-fibrotic epitenon-derived cells post-tendon injury significantly improved functional recovery. Combined, these results clearly identify the epitenon as the cellular origin of an important progenitor cell population that could be leveraged to improve tendon healing.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures







Update of
-
Epitenon-derived cells comprise a distinct progenitor population that contributes to both fibrosis and regeneration following acute flexor tendon injury in a spatially-dependent manner.bioRxiv [Preprint]. 2025 May 2:2023.01.30.526242. doi: 10.1101/2023.01.30.526242. bioRxiv. 2025. Update in: Nat Commun. 2025 Jul 1;16(1):5448. doi: 10.1038/s41467-025-60704-6. PMID: 36778469 Free PMC article. Updated. Preprint.
References
-
- Pennisi, E. Tending tender tendons. Science295, 1011 (2002). - PubMed
-
- Mehrzad, R. et al. The economic impact of flexor tendon lacerations of the hand in the United States. Ann. Plast. Surg.83, 419–423 (2019). - PubMed
-
- Strickland, J. W. & Glogovac, S. V. Digital function following flexor tendon repair in Zone II: a comparison of immobilization and controlled passive motion techniques. J. Hand Surg. Am.5, 537–543 (1980). - PubMed
-
- Quadlbauer, S. et al. Early Passive Movement in flexor tendon injuries of the hand. Arch. Orthop. Trauma. Surg.136, 285–293 (2016). - PubMed
MeSH terms
Grants and funding
- K99 AR080757/AR/NIAMS NIH HHS/United States
- R00 AR080757/AR/NIAMS NIH HHS/United States
- P30 AR069655/AR/NIAMS NIH HHS/United States
- AR077527/U.S. Department of Health & Human Services | NIH | National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS)
- AR073169/U.S. Department of Health & Human Services | NIH | National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS)
- R01 AR077527/AR/NIAMS NIH HHS/United States
- AR076950/U.S. Department of Health & Human Services | NIH | National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS)
- T32 AR076950/AR/NIAMS NIH HHS/United States
- AR080757/U.S. Department of Health & Human Services | NIH | National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS)
- R01 AR073169/AR/NIAMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

