Lysosome-mediated aggregation of galactose-deficient IgA1 with transferrin receptor 1 links to IgA nephropathy
- PMID: 40593574
- PMCID: PMC12219878
- DOI: 10.1038/s41467-025-60819-w
Lysosome-mediated aggregation of galactose-deficient IgA1 with transferrin receptor 1 links to IgA nephropathy
Abstract
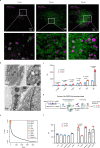
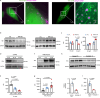
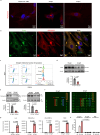
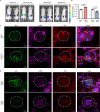
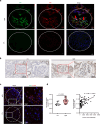
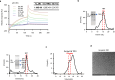
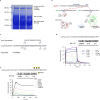
The retention of galactose-deficient IgA1 (Gd-IgA1) in the mesangium is central to IgA nephropathy (IgAN), but its intracellular fate remains unclear. Here, we show that transferrin receptor 1 (TfR1) mediates Gd-IgA1 uptake into mesangial cell lysosomes, where it forms non-digestible aggregates, disrupts lysosomal function, and triggers inflammatory responses. In renal biopsies from IgAN patients, IgA1 aggregates co-localize with TfR1 within lysosomes. In male mice, TfR1 overexpression enhanced lysosomal accumulation of Gd-IgA1, whereas TfR1 knockdown reduced it. Mechanistically, acidic pH strengthens TfR1-Gd-IgA1 binding via the galactose-deficient hinge region and residue R276. While we acknowledge that sialylation commonly found in patient-derived IgA1 might influence TfR1 binding and that other receptors, such as ASGPR, were not evaluated, our findings nonetheless reveal a lysosome-centered mechanism in IgAN and highlight receptor-mediated retention of Gd-IgA1 as a potential therapeutic target.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: All the authors declared no competing interests.
Figures


Similar articles
-
Dynamics of chromatin accessibility governing Gd-IgA1 synthesis in B cells associated with IgA nephropathy.Exp Mol Med. 2025 Jul;57(7):1593-1606. doi: 10.1038/s12276-025-01505-1. Epub 2025 Jul 23. Exp Mol Med. 2025. PMID: 40702134 Free PMC article.
-
Aberrant IgA1 Glycosylation in IgA Nephropathy: A Systematic Review.PLoS One. 2016 Nov 21;11(11):e0166700. doi: 10.1371/journal.pone.0166700. eCollection 2016. PLoS One. 2016. PMID: 27870872 Free PMC article.
-
SOCS1 improves abnormal IgA galactosylation in IgA nephropathy by regulating the TLR9/MyD88 pathway.Immunol Res. 2025 Aug 4;73(1):115. doi: 10.1007/s12026-025-09669-8. Immunol Res. 2025. PMID: 40759858
-
Glomerular galactose-deficient IgA1 and apoptosis inhibitor of macrophage staining in secondary IgA nephropathy associated with alcoholic cirrhosis.J Nephrol. 2025 Jul;38(6):1737-1742. doi: 10.1007/s40620-025-02213-9. Epub 2025 Feb 13. J Nephrol. 2025. PMID: 39946052
-
O-glycosylation of IgA1 and the pathogenesis of an autoimmune disease IgA nephropathy.Glycobiology. 2024 Sep 30;34(11):cwae060. doi: 10.1093/glycob/cwae060. Glycobiology. 2024. PMID: 39095059 Free PMC article. Review.
References
-
- Stamellou, E. et al. IgA nephropathy. Nat. Rev. Dis. Prim.9, 67 (2023). - PubMed
-
- Lai, K. N. et al. IgA nephropathy. Nat. Rev. Dis. Prim.2, 16001 (2016). - PubMed
-
- Suzuki, H. & Novak, J. IgA glycosylation and immune complex formation in IgAN. Semin. Immunopathol.43, 669–678 (2021). - PubMed
-
- Si, M. et al. Anomalous kinetics of galactose-deficient IgA incurring nephropathy revealed by cross-scale optical imaging. Kidney Int. 103, 320–330 (2023). - PubMed
-
- Sinniah, R. & Churg, J. Effect of IgA deposits on the glomerular mesangium in Berger’s disease. Ultrastruct. Pathol.4, 9–22 (1983). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

