MORC2 is a phosphorylation-dependent DNA compaction machine
- PMID: 40593625
- PMCID: PMC12216690
- DOI: 10.1038/s41467-025-60751-z
MORC2 is a phosphorylation-dependent DNA compaction machine
Abstract
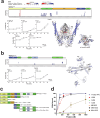
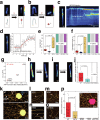
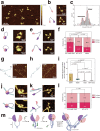
The Microrchidia (MORC) family of chromatin-remodelling ATPases is pivotal in forming higher-order chromatin structures that suppress transcription. The exact mechanisms of MORC-induced chromatin remodelling have been elusive. Here, we report an in vitro reconstitution of full-length MORC2, the most commonly mutated MORC member, linked to various cancers and neurological disorders. MORC2 possesses multiple DNA-binding sites that undergo structural rearrangement upon DNA binding. MORC2 locks onto the DNA using its C-terminal domain (CTD) and acts as a clamp. A conserved phosphate-interacting motif within the CTD was found to regulate ATP hydrolysis and cooperative DNA binding. Importantly, MORC2 mediates chromatin remodelling via ATP hydrolysis-dependent DNA compaction in vitro, regulated by the phosphorylation state of its CTD. These findings position MORC2 CTD phosphorylation as a critical regulator of chromatin remodelling and a promising therapeutic target.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

