Adipocyte-specific Zeb1 downregulation remodels the tumor-associated adipose microenvironment to facilitate female breast cancer progression
- PMID: 40593646
- PMCID: PMC12219764
- DOI: 10.1038/s41467-025-61088-3
Adipocyte-specific Zeb1 downregulation remodels the tumor-associated adipose microenvironment to facilitate female breast cancer progression
Abstract
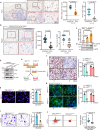
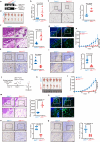
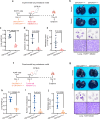
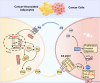
Upon penetrating the basement membrane, breast cancer cells directly interact with their surrounding adipose tissue, which forms a unique tumor-associated adipose microenvironment (TAME). However, the underlying mechanism of lipid metabolic remodeling in the TAME remains elusive. Herein, we report a Zeb1-orchestrated bidirectional communication between breast cancer cells and their adjacent cancer-associated adipocytes (CAAs). At the molecular level, breast cancer cells, through the secretion of adrenomedullin (AM), induce downregulation of Zeb1 expression to activate the Atgl/Hsl/Scd-dependent lipolysis in CAAs, resulting in the release of palmitoleic acid (POA) into the TAME. In turn, the increased POA in breast cancer competes with arachidonic acid (ARA) for the phospholipid synthesis, leaving more ARA is utilized for PDG2 production to trigger the malignant progression of breast cancer and AM production. Importantly, disruption of Zeb1-dependent lipolytic activity and/or membrane phospholipid remodeling within the TAME dramatically diminishes the aggressiveness of breast cancer in vitro and in vivo.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures





References
-
- Kong, L. R. et al. A glycolytic metabolite bypasses “two-hit” tumor suppression by BRCA2. Cell187, 2269–2287.e16 (2024). - PubMed
-
- Zhang, Y. et al. Metabolic switch regulates lineage plasticity and induces synthetic lethality in triple-negative breast cancer. Cell Metab.36, 193–208.e8 (2024). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

