Highly ordered clustering of TNFα and BAFF ligand-receptor-intracellular adaptor complexes on a lipid membrane
- PMID: 40593711
- PMCID: PMC12216653
- DOI: 10.1038/s41467-025-61271-6
Highly ordered clustering of TNFα and BAFF ligand-receptor-intracellular adaptor complexes on a lipid membrane
Abstract
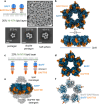
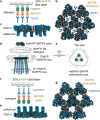
The TNF family plays a critical role in immune regulation. Here, we present high-resolution structures of clusters formed by two TNF receptor family proteins, TNFR1 and BAFFR. Using a lipid monolayer method to mimic their membrane-bound state, we observe that the TNFα-TNFR1 complex forms highly ordered clusters of trimers on the lipid membrane. A non-competitive TNFR1 antagonist that inhibits receptor activation disrupted these clusters without blocking ligand binding or receptor trimerization. Furthermore, we find that the BAFF-BAFFR, BAFF-TACI, and BAFF-BCMA receptor-ligand complexes predominantly form pentagonal clusters of trimers on the lipid membrane. Notably, the binding of the intracellular adaptor TRAF3 to the BAFF-BAFFR complex induces a structural transition from a pentagonal to a flat hexagonal cluster. Mutations in BAFF that impair BAFFR activation prevented cluster formation. Our findings demonstrate that ligand binding induces the formation of highly ordered clusters of TNFR1 and BAFFR receptors on the lipid membrane, which is essential for their activation.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures







Similar articles
-
Stoichiometry of Heteromeric BAFF and APRIL Cytokines Dictates Their Receptor Binding and Signaling Properties.J Biol Chem. 2015 Jun 26;290(26):16330-42. doi: 10.1074/jbc.M115.661405. Epub 2015 May 7. J Biol Chem. 2015. PMID: 25953898 Free PMC article.
-
No evidence that soluble TACI induces signalling via membrane-expressed BAFF and APRIL in myeloid cells.PLoS One. 2013 Apr 19;8(4):e61350. doi: 10.1371/journal.pone.0061350. Print 2013. PLoS One. 2013. PMID: 23620746 Free PMC article.
-
Selectivity of BAFF/BLyS and APRIL for binding to the TNF family receptors BAFFR/BR3 and BCMA.Biochemistry. 2005 Feb 15;44(6):1919-31. doi: 10.1021/bi048227k. Biochemistry. 2005. PMID: 15697217
-
BAFF receptor and TACI in B-1b cell maintenance and antibacterial responses.Ann N Y Acad Sci. 2015 Dec;1362:57-67. doi: 10.1111/nyas.12772. Epub 2015 May 11. Ann N Y Acad Sci. 2015. PMID: 25962322 Review.
-
BAFF, APRIL and their receptors: structure, function and signaling.Semin Immunol. 2006 Oct;18(5):263-75. doi: 10.1016/j.smim.2006.04.006. Epub 2006 Aug 17. Semin Immunol. 2006. PMID: 16914324 Review.
References
-
- Dostert, C., Grusdat, M., Letellier, E. & Brenner, D. The TNF family of ligands and receptors: communication modules in the immune system and beyond. Physiol. Rev.99, 115–160 (2019). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

