Evolutionary regulation of human Fas ligand (CD95L) by plasmin in solid cancer immunotherapy
- PMID: 40593750
- PMCID: PMC12217004
- DOI: 10.1038/s41467-025-60990-0
Evolutionary regulation of human Fas ligand (CD95L) by plasmin in solid cancer immunotherapy
Abstract
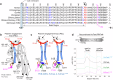
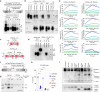
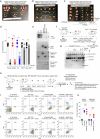
Despite sharing >98% genomic similarity, humans are more likely to develop cancers than our closest living ancestors, the nonhuman primates. Here, we unexpectedly discover that, unlike chimpanzee and other primates, a critical embryonic development, immune homeostasis, and general cell-death regulator protein called Fas Ligand (FasL) contains a Pro153-Ser153 evolutionary substitution in humans. The latter renders human FasL preferentially susceptible to cleavage by plasmin, an overly elevated protease in solid tumors. Since FasL-mediated killing of tumor cells by activated T-lymphocytes and chimeric antigen receptor T-cells (CAR-T) is critical for therapeutic efficacy, we find that elevated plasmin levels in certain ovarian tumors interfere with the T-lymphocyte-expressed FasL death signaling. Either targeted inhibition or blocking plasmin accessibility to membrane FasL rescues the FasL cell-death function of activated T-lymphocytes in response to immune-checkpoint receptor targeting antibodies. These findings of evolutionary significance highlight that elevated plasmin in metastatic tumors potentially contributes to differential outcomes of T-cell-based immunotherapies in solid tumors.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures






References
-
- Kelly, P. N. The cancer immunotherapy revolution. Science359, 1344–1345 (2018). - PubMed
-
- Coulie, P. G., Van den Eynde, B. J., van der Bruggen, P. & Boon, T. Tumour antigens recognized by T lymphocytes: at the core of cancer immunotherapy. Nat. Rev. Cancer14, 135–146 (2014). - PubMed
-
- Peggs, K. S., Quezada, S. A. & Allison, J. P. Cell intrinsic mechanisms of T-cell inhibition and application to cancer therapy. Immunol. Rev.224, 141–165 (2008). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

