Characterizing organisms from three domains of life with universal primers from throughout the global ocean
- PMID: 40593763
- PMCID: PMC12218243
- DOI: 10.1038/s41597-025-05423-9
Characterizing organisms from three domains of life with universal primers from throughout the global ocean
Abstract
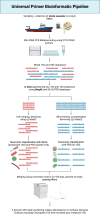
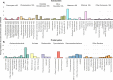
We introduce the Global rRNA Universal Metabarcoding Plankton database (GRUMP), which consists of 1194 samples that were collected from 2003-2020 and cover extensive latitudinal and longitudinal transects, as well as depth profiles in all major ocean basins. DNA from unfractionated (>0.2 µm) seawater samples was amplified using the 515Y/926 R universal three-domain rRNA gene primers, simultaneously quantifying the relative abundance of amplicon sequencing variants (ASVs) from bacteria, archaea, eukaryotic nuclear 18S, and eukaryotic plastid 16S. Thus, the ratio between taxa in one sample is directly comparable to the ratio in any other GRUMP sample, regardless of gene copy number differences. This obviates a problem in prior global studies that used size-fractionation and different rRNA gene primers for bacteria, archaea, and eukaryotes, precluding comparisons across size fractions or domains. On average, bacteria contributed 71%, eukaryotes 19%, and archaea 8% to rRNA gene abundance, though eukaryotes contributed 32% at latitudes >40°. GRUMP is publicly available on the Simons Collaborative Marine Atlas Project (CMAP), promoting the global comparison of marine microbial dynamics.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



References
-
- Azam, F. et al. The Ecological Role of Water-Column Microbes in the Sea. Mar. Ecol. Prog. Ser.10, 257–263 (1983). - DOI
-
- Steele, J. H. The Structure of Marine Ecosystems. 10.4159/harvard.9780674592513 (Harvard University Press, 2013).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

