Lactate promotes invasive Klebsiella pneumoniae liver abscess syndrome by increasing capsular polysaccharide biosynthesis via the PTS-CRP axis
- PMID: 40593854
- PMCID: PMC12216495
- DOI: 10.1038/s41467-025-61379-9
Lactate promotes invasive Klebsiella pneumoniae liver abscess syndrome by increasing capsular polysaccharide biosynthesis via the PTS-CRP axis
Abstract
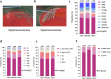
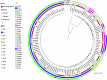
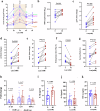
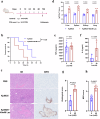
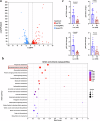
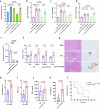
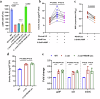
The global incidence of invasive Klebsiella pneumoniae liver abscess syndrome (IKPLAS) increases, yet its underlying molecular mechanisms remain elusive, hindering the development of effective therapeutic strategies. In this study, we analyze bacterial molecular profiles and clinical data from patients with KPLA and IKPLAS, and find no significant difference in the molecular characteristics of K. pneumoniae between the two groups, however, we identify elevated blood lactate levels as an independent predictor of IKPLAS. Further investigation reveals that lactate enhances K. pneumoniae virulence by promoting capsular polysaccharide (CPS) biosynthesis. Mechanistically, lactate reduces cyclic adenosine monophosphate (cAMP) levels by downregulating the expression of mannose-specific phosphotransferase system (man-PTS) enzyme IIA-D genes (gfrA, gfrB, gfrC and gfrD). This reduction in cAMP levels enhances CPS biosynthesis by decreasing its binding to the cAMP receptor protein (CRP). Our results highlight lactate's role in enhancing the virulence of K. pneumoniae via the PTS-CRP axis, offering insights into the pathogenesis of IKPLAS.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures
Similar articles
-
Involvement of cAMP receptor protein in biofilm formation, fimbria production, capsular polysaccharide biosynthesis and lethality in mouse of Klebsiella pneumoniae serotype K1 causing pyogenic liver abscess.J Med Microbiol. 2017 Jan;66(1):1-7. doi: 10.1099/jmm.0.000391. Epub 2017 Feb 6. J Med Microbiol. 2017. PMID: 27902401
-
Type I-E* CRISPR-Cas of Klebsiella pneumoniae upregulates bacterial virulence by targeting endogenous histidine utilization system.mSphere. 2025 Jun 25;10(6):e0021525. doi: 10.1128/msphere.00215-25. Epub 2025 May 19. mSphere. 2025. PMID: 40387367 Free PMC article.
-
Evaluation of capsule polysaccharide (CPS)-specific antibodies for broad recognition of prominent multidrug-resistant Klebsiella pneumoniae.Microbiol Spectr. 2025 Jul;13(7):e0333824. doi: 10.1128/spectrum.03338-24. Epub 2025 May 22. Microbiol Spectr. 2025. PMID: 40401961 Free PMC article.
-
Klebsiella pneumoniae capsular polysaccharide: Mechanism in regulation of synthesis, virulence, and pathogenicity.Virulence. 2024 Dec;15(1):2439509. doi: 10.1080/21505594.2024.2439509. Epub 2024 Dec 13. Virulence. 2024. PMID: 39668724 Free PMC article. Review.
-
New insights and perspectives on the virulence of hypervirulent Klebsiella pneumoniae.Folia Microbiol (Praha). 2025 Jun;70(3):517-533. doi: 10.1007/s12223-025-01261-9. Epub 2025 Apr 8. Folia Microbiol (Praha). 2025. PMID: 40198504 Review.
References
-
- Siu, L. K., Yeh, K.-M., Lin, J.-C., Fung, C.-P. & Chang, F.-Y. Klebsiella pneumoniae liver abscess: a new invasive syndrome. Lancet Infect. Dis.12, 881–887 (2012). - PubMed
-
- Zhang, J. et al. Clinical features and prognosis of gas-forming and non-gas-forming pyogenic liver abscess: a comparative study. Surg. Infect. (Larchmt.)22, 427–433 (2021). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

