Histone lysine methyltransferases MLL3 and MLL4 direct gene expression to produce platelets efficiently
- PMID: 40593910
- PMCID: PMC12215508
- DOI: 10.1038/s41467-025-61247-6
Histone lysine methyltransferases MLL3 and MLL4 direct gene expression to produce platelets efficiently
Abstract
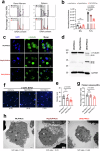
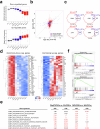
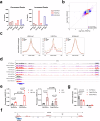
Circulating blood platelets are responsible for maintaining hemostasis. They are released into blood vessels from mature megakaryocytes. Although several transcription factors have been reported to orchestrate the transcriptional programs required for platelet production, how chromatin regulators control these processes is still poorly understood. MLL3 and MLL4 are the main lysine methyltransferases responsible for the deposition of H3K4me1 histone marks at enhancers. MLL3 and MLL4 typically form complexes with other co-factors, such as PTIP. Recently, we showed that loss of PTIP leads to decreased platelet numbers in mice. Here, we find that, although MLL3/4 double deficiency does not alter megakaryopoiesis and endomitosis, the final step of megakaryocyte maturation is affected due to an abnormal cytoskeleton and demarcation membrane system. MLL3/4 double-deficient mice develop macrothrombocytopenia; platelets are preactive and pro-apoptotic, leading to their rapid clearance from the circulation. Increased megakaryopoeisis in the bone marrow and spleen cannot compensate for these abnormalities. Mechanistically, the expression of genes responsible for normal megakaryocyte function and platelet production is altered in MLL3/4-deficient megakaryocytes, partly due to impaired enhancer functions associated with these genes. Our findings provide insights into the epigenetic programs that are important for platelet biogenesis.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare that they have no conflict of interest.
Figures




References
-
- Eckly, A. et al. Biogenesis of the demarcation membrane system (DMS) in megakaryocytes. Blood123, 921–930 (2014). - PubMed
-
- Mitchell, W. B. & Bussel, J. B. Thrombopoietin receptor agonists: a critical review. Semin Hematol.52, 46–52 (2015). - PubMed
-
- Hart, A. et al. Fli-1 is required for murine vascular and megakaryocytic development and is hemizygously deleted in patients with thrombocytopenia. Immunity13, 167–177 (2000). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

