Identification of aging-related biomarkers and immune infiltration analysis in renal stones by integrated bioinformatics analysis
- PMID: 40593989
- PMCID: PMC12214660
- DOI: 10.1038/s41598-025-05087-w
Identification of aging-related biomarkers and immune infiltration analysis in renal stones by integrated bioinformatics analysis
Abstract
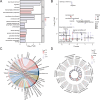
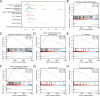
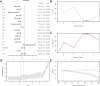
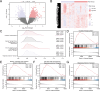
Renal stones (RS) are common urologic condition with unclear pathogenesis. Role of aging-related differentially expressed genes (ARDEGs) in RS remains poorly understood. This study aims to identify potential aging-related biomarkers for RS, explore the functions of aging-associated genes, and investigate the immunological microenvironment in RS. ARDEGs were collected from the GEO, GeneCards, and Molecular Signatures databases. The roles of ARDEGs were analyzed using Gene Ontology (GO) enrichment analysis. Key genes were identified using machine learning methods. Immune infiltration in RS was assessed using the CIBERSORT and ssGSEA algorithms. A total of 22 ARDEGs were identified through analysis, including 9 up-regulated and 13 down-regulated genes. GO enrichment analysis revealed that these genes were mainly involved in RS-related biological processes such as macrophage proliferation and neuroinflammatory response. GSEA analysis showed that RS-associated genes were predominantly involved in immune regulation-related pathways. Using logistic regression, SVM, and LASSO regression algorithms, a successful early-diagnosis model for RS was developed, yielding 7 key genes: CNR1, KIT, HTR2A, DES, IL33, UCP2, and PPT1. Immunocyte infiltration analysis of RS samples showed that CD8 + T cells had the strongest positive correlation with M1 macrophages, while resting NK cells had the strongest negative correlation with activated NK cells. The DES gene showed the strongest positive correlation with resting mast cells, and the IL33 gene displayed the highest negative correlation with regulatory T cells. Bioinformatics analysis screened out 7 new potential markers for RS and explored the possible mechanism of RS senescence. These findings provide novel insights into the relationship between RS and senescence, as well as the diagnosis and treatment of RS, and enhance our understanding of the disease's occurrence and development mechanisms.
Keywords: Biomarkers; Immune Infiltration; Integrated Bioinformatics; Renal Stone.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures







References
-
- Thongprayoon, C., Krambeck, A. E. & Rule, A. D. Determining the true burden of kidney stone disease. Nat. Rev. Nephrol.16(12), 736–746 (2020). - PubMed
-
- Lyall, V. S., Wood, K. D. & Pais, V. J. Hydrochlorothiazide and Prevention of Kidney-Stone Recurrence. N. Engl. J. Med.388(21), 2014 (2023). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

