Complete genomic characterization of global pathogens respiratory syntical virus and human norovirus using probe based capture enrichment
- PMID: 40594045
- PMCID: PMC12216758
- DOI: 10.1038/s41598-025-03398-6
Complete genomic characterization of global pathogens respiratory syntical virus and human norovirus using probe based capture enrichment
Abstract
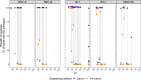
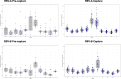
Respiratory syncytial virus (RSV) is the leading cause of lower respiratory tract infections in children worldwide, while human noroviruses (HuNoV) are a leading cause of epidemic and sporadic acute gastroenteritis. Generating full-length genome sequences for these viruses is crucial for understanding viral diversity and tracking emerging variants. However, obtaining high-quality sequencing data is often challenging due to viral strain variability, quality, and low titers. Here, we present a set of comprehensive oligonucleotide probe sets designed from 1,570 RSV and 1,376 HuNoV isolate sequences in GenBank. Using these probe sets and a capture enrichment sequencing workflow, 85 RSV positive nasal swab samples and 55 (49 stool and six human intestinal enteroids) HuNoV positive samples encompassing major subtypes and genotypes were characterized. Samples with Ct values 17.0-29.9 for RSV, and 20.2-34.8 for HuNoV, with some HuNoV below the detection limit were sequenced. The percentage of reads mapped to viral genomes was 85.1% for RSV and 40.8% for HuNoV post-capture, compared to 0.08% and 1.15% in pre-capture libraries. Full-length genomes were obtained for all RSV positive samples and in 47/55 HuNoV positive samples-a significant improvement over genome recovery from pre-capture libraries. RSV transcriptome (subgenomic mRNAs) sequences were also characterized from this data.
Keywords: Capture enrichment; Genome sequencing; Human norovirus (HuNoV); Respiratory syncytial virus (RSV).
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: R.L.A. and M.K.E. have grant support from Hillevax, Inc. and are consultants for that company. Baylor College of Medicine (R.L.A. and M.K.E. as inventors) has a patent for norovirus growth in human intestinal enteroids, and M.K.E. has a patent on methods and reagents to detect and characterize Norwalk virus and related viruses. The rest of the authors declare no competing interests. Consent to participate: All methods were performed in accordance with relevant guidelines and regulations. Ethical approval: RSV-positive samples were collected from patients enrolled at the Houston NVSN site only. They were obtained after written informed consent was obtained from the parent/guardian of the eligible children and age-appropriate assent from participating children. Institutional review board approval was obtained locally from Baylor College of Medicine (H-37691) and at the CDC. Norovirus-positive samples were collected as deidentified samples prior to disposal from a clinical laboratory or as residual samples collected in a clinical study. The latter samples were obtained after written informed consent was obtained from the participants. Institutional review board approval was obtained locally from Baylor College of Medicine for both study groups (H-8390, H-45026).
Figures



Update of
-
Complete Genomic Characterization of Global Pathogens, Respiratory Syncytial Virus (RSV), and Human Norovirus (HuNoV) Using Probe-based Capture Enrichment.bioRxiv [Preprint]. 2024 Sep 16:2024.09.16.613242. doi: 10.1101/2024.09.16.613242. bioRxiv. 2024. Update in: Sci Rep. 2025 Jul 1;15(1):20526. doi: 10.1038/s41598-025-03398-6. PMID: 39345650 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

